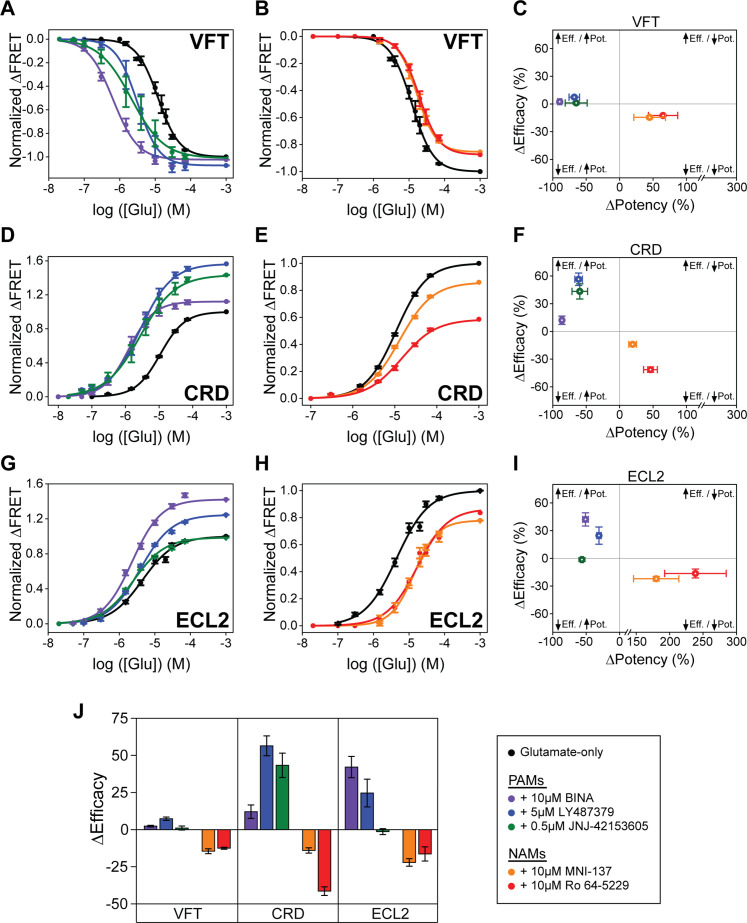
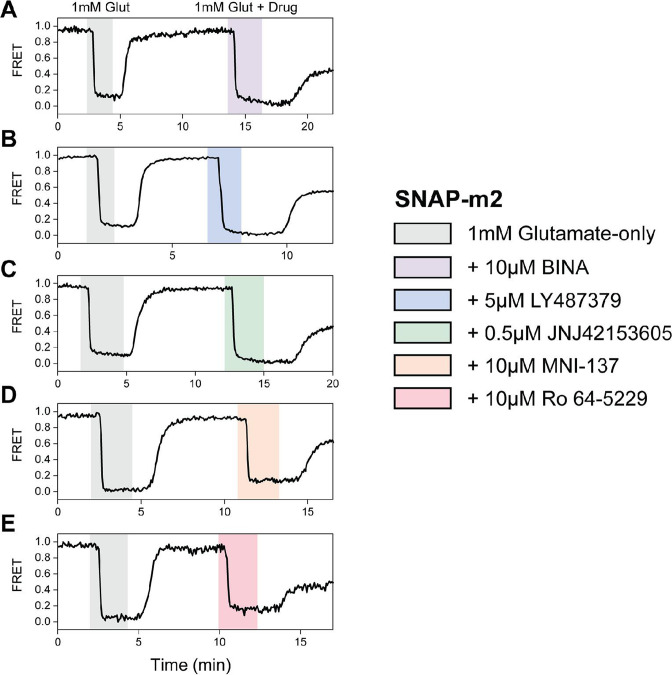
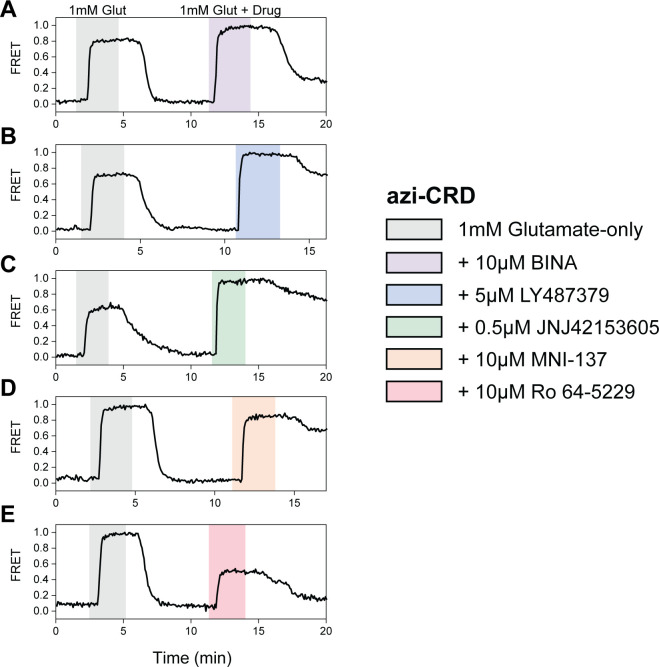
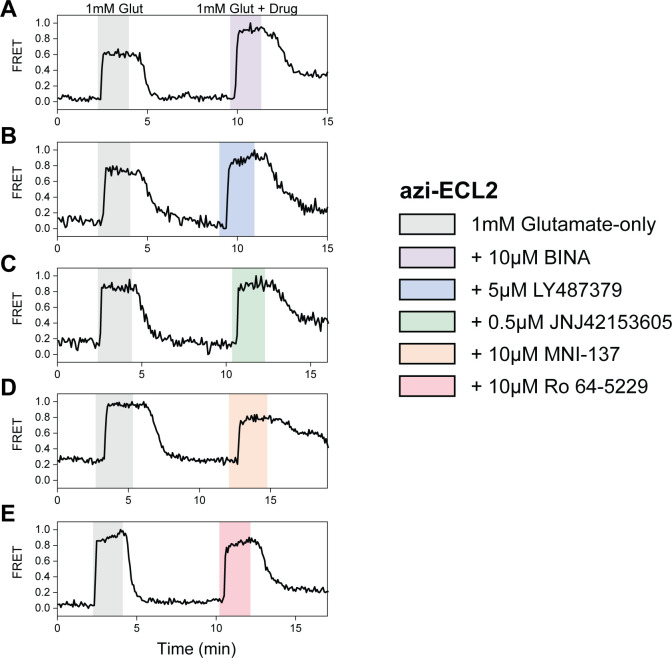
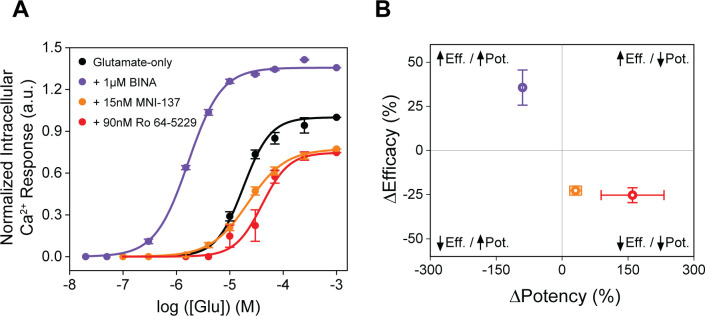
Figure 2. Positive and negative allosteric modulation of metabotropic glutamate receptor 2 (mGluR2) structural domains.
N-terminal SNAP-tag labeled mGluR2; hereafter (SNAP-m2) glutamate dose-response curves in the presence of (A) positive allosteric modulators (PAMs) or (B) NAMs. (C) Changes in glutamate potency and efficacy for SNAP-m2. The azi-cysteine-rich domain (azi-CRD) glutamate dose-response curves in the presence of (D) PAMs or (E) NAMs. (F) Changes in glutamate potency and efficacy for azi-CRD. The azi-extracellular loop 2 (azi-ECL2) glutamate dose-response curves in the presence of (G) PAMs or (H) NAMs. (I) Changes in glutamate potency and efficacy for azi-ECL2. (J) Changes in glutamate efficacy in response to PAMs and NAMs as measured by each conformational sensor. ΔPotency defined as (([modulator + glutamate]EC50 – [glutamate] EC50)/[glutamate] EC50) × 100. ΔEfficacy defined as ([1 mM glutamate + modulator] – [1 mM glutamate]) × 100. Data is acquired from individual cells and normalized to 1 mM glutamate response. Data represents mean ± SEM of responses from individual cells from at least three independent experiments. Total number of cells examined for titration and normalization experiments, mean half-maximum effective concentration (EC50), mean max response, and errors are listed in Tables 1–2.