Abstract
Purpose:
To determine the degree of testing consistency among commercially available diagnostic assays for hereditary hematopoietic malignancies (HHMs).
Methods:
Next-generation sequencing assays designed for the diagnosis of HHMs were studied to determine which genes were sequenced, their ability to detect variant types relevant for HHMs, and clinical-grade characteristics such as price, turnaround time, and tissue types accepted.
Results:
Commercial assays varied in price (USD 250–4702), number of genes sequenced (12–73), and average turnaround time (14–42 days). A number of nongermline tissue types were accepted despite the tests being designed for germline diagnostic purposes. Multiple genes with well-characterized roles in HHM pathogenesis were omitted from more than one-third of panels intended for the evaluation of HHMs. Only 4 of 82 genes were consistently covered across all HHM diagnostic panels. The assays were highly variable in their sensitivity for structural alterations relevant to HHMs, such as copy-number variants.
Conclusion:
A high degree of diagnostic heterogeneity exists among commercially available HHM diagnostic assays. Many of these assays are incapable of detecting the full spectrum of HHM-associated variants, leaving patients vulnerable to the consequences of underdiagnosis, missed opportunities for screening, and the potential for donor-derived malignancies.
Keywords: hematopoietic malignancies, germline, panel testing, inherited leukemia, cancer risk
INTRODUCTION
Hereditary hematopoietic malignancies (HHMs) are syndromes driven by germline variants that substantially increase an individual’s lifetime risk of hematopoietic malignancies.1 HHMs follow Mendelian inheritance patterns, and multiple individuals within a single family are often affected.1 Although HHMs have been described for decades, recent advances in next-generation sequencing (NGS) technologies have accelerated the rate of HHM discovery. Germline variants in more than a dozen genes are now recognized as HHM drivers.1,2
Patients with HHMs may be identified when “somatic” tumor NGS panels incidentally detect germline variants passed into the malignant clone.3 Several laboratories now market assays designed specifically for the diagnosis of HHMs. However, we have cared for multiple patients for whom initial HHM testing was not sensitive either to the nature of the potential germline variant or the relevant genomic locus. These individuals were found eventually to carry likely pathogenic or pathogenic HHM-associated variants that were missed by the initial assay. For example, we have encountered patients with HHMs and personal/family histories of thrombocytopenia and/or excessive bleeding for whom commercial assays failed to include the 5’ untranslated region of ANKRD26, the mutational hotspot for HHMs driven by germline ANKRD26 variants.4 In a separate case, a referring physician for an individual with a similar personal/family history ordered an NGS panel that covered only point variants in RUNX1, failing to order the test with automatic reflexing to a larger panel that included ANKRD26 and ETV6, as well as an approach that could detect copy-number variants (CNVs) in all three genes. Comprehensive testing subsequently identified a multiexon RUNX1 deletion. We have met other patients with active hematopoietic malignancies who had germline testing performed on DNA derived from peripheral blood samples that contained tumor cells.
These examples highlight the need for reliable and comprehensive testing standards for the evaluation of HHMs, including coverage for the wide variety of known variant types (i.e., point variants/single-nucleotide variants [SNVs] and CNVs). A systematic analysis of testing methods among commercial laboratories, however, has never been performed in the field of HHMs. Therefore, we hypothesized that substantial technical variability exists among commercially available assays and that the ability to detect HHM-associated germline variants accurately varies from laboratory to laboratory.
MATERIALS AND METHODS
We evaluated commercially available NGS assays marketed specifically for the evaluation of HHMs. We analyzed the websites of established genetics laboratories and queried the US National Center for Biotechnology Information’s (NCBI) Genetic Testing Registry (https://www.ncbi.nlm.nih.gov/gtr/) to identify assays intended for use in suspected HHMs, specifically hereditary myelodysplastic syndromes and acute leukemia (MDS/AL) (n = 8). Data were also collected for inherited bone marrow failure syndrome (IBMFS) testing, which can be found in the Supplementary Materials (n = 8). We compared the number of genes included in each test, cost, turnaround time, specimen types accepted, and sequencing-specific variables, such as resolution of CNV detection and methodologies for confirmation of SNVs. We excluded NGS panels intended for somatic variant profiling from our analyses. As assays from some laboratories are updated regularly, we performed our final analyses on 2 May 2020. We contacted each laboratory and provided them with ten business days to review, clarify, or contest any data regarding the attributes or performance of their assays. Ninety percent of the laboratories responded to this query.
RESULTS
An overview of assay characteristics and select sequencing attributes is displayed in Table 1. Commercial assays varied in price (ranging from USD250 to 4702), number of genes sequenced (12 to 73), and average turnaround time (14 to 42 days). There was substantial variability in terms of tissue types accepted for analysis, and many laboratories accepted peripheral blood as a tissue specimen. Blood is a particularly inappropriate source of DNA for the diagnosis of some HHMs. For example, HHMs driven by pathogenic germline SAMD9/SAMD9L variants are notable for genetic reversion events that are difficult to detect via sequencing of hematopoietic tissue alone.5 Overall, 63% of the laboratories in this study sequenced SAMD9/SAMD9L. Of these laboratories, 40% accepted peripheral blood either as a sole source of DNA or did not include language that emphasized the importance of sequencing nonhematopoietic tissue for the accurate diagnosis of HHMs driven by pathogenic germline variants in SAMD9/SAMD9L. Peripheral blood may also contain variants associated with clonal hematopoiesis (CH) even when complete morphological remission is achieved, and these variants may be confused for HHMs.6 For example, persistent TP53 variants observed in CH may be mistaken for Li–Fraumeni syndrome.7 Some laboratories issued disclaimers about the importance of submitting nonblood samples when conducting germline testing for HHMs (Table 1). However, these disclaimers were not always readily apparent on company websites and were only discovered upon review of supplemental documents. A minority of laboratories reported that they would notify ordering physicians who provided nongermline tissue, but these laboratories will run the sample at the discretion of the ordering physician or if alternative tissue is not available.
Table 1.
Practical and technical attributes of commercial HHM assays.
| Company/institution | Preferred Specimen | # Genes included | List price (USD) | Turnaround (days) | CNV resolution/limitations | CNV confirmation | SNV confirmation |
|---|---|---|---|---|---|---|---|
| Laboratory A | WBa | 16 | 250b | 14 | Single-exon resolution | MLPA | Long-read sequencing |
| Laboratory B | SF | 12 | 990 | 18 | ~80% sensitivity for CNVs <4 exons; ~100% sensitivity for CNVs >4 exons | aCGH, MLPA | Upon review |
| Laboratory C | WB, purified DNA, salivaa | 41 | 1600 | 28 | May not reliably detect single-exon CNVs or indels >50 bp | ddPCR if <10 exons | Upon review |
| Laboratory D | SF | 12 | 3285 | 21 | Cannot reliably detect 20–250 bp deletions or 10–250 bp insertions | aCGH, MLPA | Upon review |
| Laboratory E | WB, saliva, buccala | 16 | 1450 | 14–21 | Single-exon resolution | MLPA, qPCR | Sanger |
| Laboratory F | SF | 28 | 4000 | 42 | May not reliably detect partial-exon CNVs or rearrangements <400 bp | MLPA, qPCR | Sanger |
| Laboratory G | WBa | 12 | 3500 | 28 | Single-exon resolution | aCGH, MLPA | Sanger |
| Laboratory H | WB | 73 | 4702 | 42 | Reliably detects CNVs of 3+ exons | MLPA, ddPCR | Sanger |
Eight HHM assays were identified. Data were collected from laboratory websites, test requisition forms, and test information sheets. Laboratory representatives were contacted to verify the data. Numerous tissue specimen types were accepted. “Upon review” indicates that variants are not reflexively validated but are instead confirmed by secondary methodology only if internal quality standards are not met. Commercial assays analyzed: Baylor Genetics Hereditary Leukemia/Lymphoma panel, Blueprint Genetics Hereditary Leukemia Panel, University of Chicago Medical Center Familial Myelodysplastic Syndrome/Acute Leukemia Panel, Fulgent Hematologic Malignancy Comprehensive Panel, GeneDx Hereditary MDS/Leukemia Panel, Invitae Myelodysplastic Syndrome/Leukemia Panel, Children’s Hospital of Philadelphia Hereditary MDS/Leukemia Predisposition Panel, and Prevention Genetics Hereditary Myelodysplastic Syndrome (MDS)/Acute Myeloid Leukemia (AML) Panel. Genes included reflect those on primary MDS/AL HHM panels for each laboratory and excluded “add-on” genes.
aCGH array comparative genomic hybridization, CNV copy-number variant, ddPCR droplet digital polymerase chain reaction, HHM hereditary hematopoietic malignancies, indel insertion/deletion, MLPA multiplex ligation-dependent probe amplification, qPCR quantitative polymerase chain reaction, SF skin fibroblasts, SNV singlenucleotide variant, WB whole blood.
Some laboratories indicated the need for nonblood specimens in patients with active hematopoietic malignancies or who had received allogenic transplants.
Price reflects the list price before the application of health insurance cost reductions or maximum out-of-pocket (b) policies adopted by some entities.
Next, we evaluated the commercial assays for inclusion of HHM-associated genes. To this end, we employed a binary matrix approach wherein any single gene included on one panel was cross-referenced for inclusion on all other assays. We found marked discordance among testing platforms, as only 4 of 82 total genes (CEBPA, GATA2, RUNX1, TP53) were included on all MDS/AL panels. Given the large number of genes that appeared across all commercial assays, we focused the next phase of our analysis on a subset of genes that appeared on at least half (4/8) of all MDS/AL panels (n = 25) (Fig. 1). Despite their well-characterized role in causing HHMs, ANKRD26, ETV6, SAMD9, and SAMD9L were not included on 3/8 panels (~37%) (Fig. 1). We also noted substantial inconsistency across IBMFS testing, which sequenced a total of 212 genes across all assays, but only covered a minority of genes consistently (BRCA2, select Fanconi anemia genes, GATA1, GATA2, and TP53) (Supplementary Materials).
Fig. 1. Genes sequenced on commercial panels for hereditary hematopoietic malignancies.
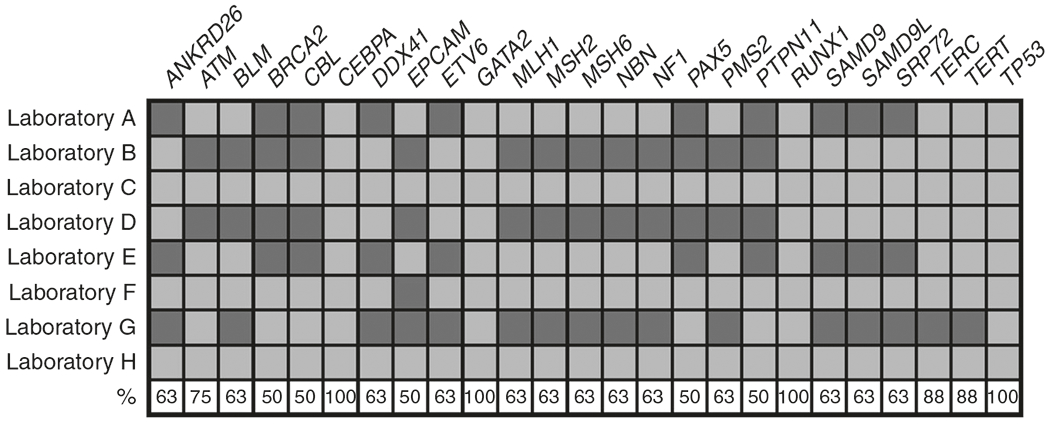
The matrix demonstrates if each gene of interest was analyzed by each individual laboratory. Percentage of laboratories sequencing each gene of interest is shown as a percent in the bottom row. Only genes included on at least half (4/8) of panels are included. Orange: gene analyzed; blue: gene omitted.
The genes sequenced by each laboratory did not necessary correlate with the importance of these genes in HHM pathogenesis. For example, germline variants in DDX41 are likely the most prevalent germline genetic driver of HHMs.8,9 Approximately 1% of all seemingly sporadic cases of acute leukemia are driven by germline DDX41 variants,10 and ClinVar reports 18 pathogenic/likely pathogenic variants of germline or de novo origin (Supplementary Materials). Despite the significant role of DDX41 in HHMs, only 5/8 commercial assays analyzed DDX41. In contrast, EPCAM, which is not known to cause HHMs, was included on 50% of MDS/AL and 25% of IBMFS panels.
DISCUSSION
Identifying an HHM provides multiple benefits for patients and their family members.11 Since matched related donor transplants are an effective therapy for many hematologic disorders, screening of related donor candidates may reduce the likelihood of donor-derived hematopoietic malignancies.12 Donor screening may also allow unaffected HHM variant carriers to avoid the currently unclear risks associated with stem cell mobilization. Additionally, family members may undergo evaluation via cascade testing following an HHM diagnosis in a relative. Variants in many HHM-associated genes also confer solid tumor risk,11 and cascade testing may provide opportunities for screening and surveillance of other cancer types.
Our analysis demonstrates that a high degree of diagnostic heterogeneity exists among commercially available HHM assays. Many of these commercial assays are not capable of detecting the full spectrum of HHM-associated variants, and this deficiency may lead to the underdiagnosis of HHMs in at risk patients and ultimately to suboptimal care of these individuals, potentially increasing the risk of donor-derived leukemias.
Expert panels within ClinGen (https://clinicalgenome.org/affiliation/50034/) have been established to curate variants in HHM-associated genes,13 and continued emphasis on testing consistency will unify commercial entities toward optimized panel content and consensus diagnostic guidelines for HHM assays. These standards would detail the specific genes that must be sequenced, technical approaches that are needed to detect the full spectrum of variant types responsible for HHMs, and the tissue types that are acceptable for accurate analyses. Ultimately, these guidelines will benefit the larger HHM community of patients, researchers, and clinicians. Improving the consistency of HHM clinical variant interpretation will ultimately lead to more uniform care of populations at risk for HHMs.
Although none of the laboratories in this study indicated that they intend to transition to an unbiased sequencing approach in the short term, we expect that germline sequencing assays will utilize unbiased sequencing methods, such as exome/genome sequencing, increasingly with time as the cost of NGS declines. Genome sequencing, in particular, would also facilitate the analysis of intronic regions that are known to harbor HHM-causative variants, but that the majority of tests do not currently sequence. For example, pathogenic germline variants in noncoding regions of GATA2 drive some HHMs, but only 3 (38%) of the laboratories sequenced these intronic regions. Of note, genome sequencing alone will not be sufficient for the detection of these variants unless the sequencing is paired with a truly comprehensive analysis of all known pathogenic variants for HHMs. Therefore, these unbiased efforts will still require the careful analysis of a standard set of genomic regions that contain known pathogenic or likely pathogenic HHM-associated variants. FDA-recognized variant curation by the Myeloid Malignancy Variant Curation Expert Panel and other ClinGen panels will continue to inform these efforts, ultimately reducing the likelihood of false-negative genetic testing in patients harboring a germline variant, decreasing the risk of donor cell derived leukemias, better informing cascade testing in unaffected family members, and likely increasing enrollment in HHM research programs.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge patients and families who participate in HHM research programs. We also thank the commercial and academic laboratories mentioned above for their responsiveness and thoughtful feedback. G.W.R. is supported by the Research Scholar Track of the Loyola University Internal Medicine Residency Program. M.W.D. is supported by the Damon Runyon Cancer Research Foundation and the NIH Paul Calabresi K12 Scholars Program.
Footnotes
SUPPLEMENTARY INFORMATION
The online version of this article (https://doi.org/10.1038/s41436-020-0934-y) contains supplementary material, which is available to authorized users.
DISCLOSURE
L.A.G. serves as a scientific advisor for Invitae and receives royalties from an article about “Familial acute leukemia and myelodysplastic syndromes” for UpToDate, Inc. The other authors declare no conflicts of interest.
REFERENCES
- 1.Akpan IJ, Osman AEG, Drazer MW, Godley LA. Hereditary myelodysplastic syndrome and acute myeloid leukemia: diagnosis, questions, and controversies. Curr Hematol Malig Rep. 2018;13:426–434. [DOI] [PubMed] [Google Scholar]
- 2.Duployez N, Willekens C, Plo I, Marceau-Renaut A, de Botton S, Fenwarth L, et al. Inherited transmission of the CSF3R T618I mutational hotspot in familial chronic neutrophilic leukemia. Blood. 2019;134:2414–2416. [DOI] [PubMed] [Google Scholar]
- 3.Drazer MW, Kadri S, Sukhanova M, Patil SA, West AH, Feurstein S, et al. Prognostic tumor sequencing panels frequently identify germ line variants associated with hereditary hematopoietic malignancies. Blood Adv. 2018;2:146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noris P, Favier R, Alessi MC, Geddis AE, Kunishima S, Heller PG, et al. ANKRD26-related thrombocytopenia and myeloid malignancies. Blood. 2013;122:1987–1989. [DOI] [PubMed] [Google Scholar]
- 5.Nagata Y, Narumi S, Guan Y, Przychodzen BP, Hirsch CM, Makishima H, et al. Germline loss-of-function SAMD9 and SAMD9L alterations in adult myelodysplastic syndromes. Blood. 2018;132:2309–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35:934–946. [DOI] [PubMed] [Google Scholar]
- 7.Steensma DP. Clinical implications of clonal hematopoiesis. Mayo Clin Proc. 2018;93:1122–1130. [DOI] [PubMed] [Google Scholar]
- 8.Tawana K, Fitzgibbon J. Inherited DDX41 mutations: 11 genes and counting. Blood. 2016;127:960–961. [DOI] [PubMed] [Google Scholar]
- 9.Lewinsohn M, Brown AL, Weinel LM, Phung C, Rafidi G, Lee MK, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tawana K, Drazer MW, Churpek JE. Universal genetic testing for inherited susceptibility in children and adults with myelodysplastic syndrome and acute myeloid leukemia: are we there yet? Leukemia. 2018;32:1482–1492. [DOI] [PubMed] [Google Scholar]
- 12.Engel N, Rovo A, Badoglio M, Labopin M, Basak GW, Beguin Y, et al. European experience and risk factor analysis of donor cell-derived leukaemias/MDS following haematopoietic cell transplantation. Leukemia. 2019;33:508–517. [DOI] [PubMed] [Google Scholar]
- 13.Luo X, Feurstein S, Mohan S, Porter CC, Jackson SA, Keel S, et al. ClinGen Myeloid Malignancy Variant Curation Expert Panel recommendations for germline RUNX1 variants. Blood Adv. 2019;3: 2962–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


