Fig. 2: Culture of CMs in vitro recapitulates the in vivo phenotype, and further shows that substrate stiffness disrupts nuclear organization and the expression of histones and histone modifying enzymes.
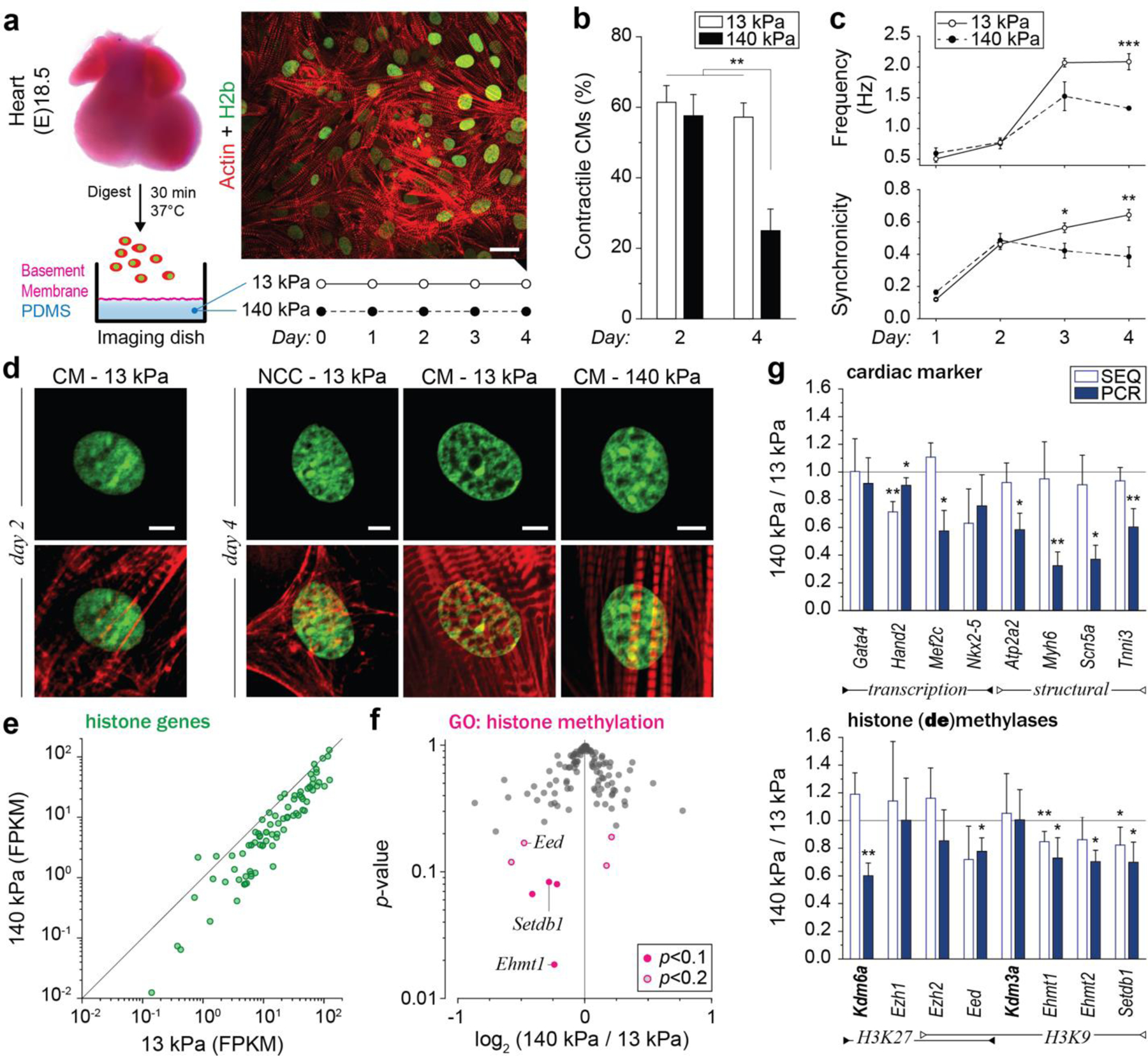
a) A warm digestion protocol was established using ECM-specific peptidases to isolate cells from (E)18.5 H2b-eGFP embryo hearts with high ratios of CMs. Cardiac cells were cultured on soft (13 kPa, shown) or stiff (140 kPa) Geltrex-coated PDMS substrates; scale=20 µm. b) After two and four days, cultures were stained for actin and the ratio of CMs with contractile myofibrils to non-contractile cells (NCC) was determined. The percentage of contractile CMs was significantly reduced after four days on stiff substrates compared to soft. SEM, n=9 from 3 exp., 2W-ANOVA: ** p<0.01. c) CMs were stained with Fluo-4 to analyze Ca2+-signaling. Cells on stiff substrates showed reduced frequency and synchronicity after 4 days in culture compared to cells on soft. d) Embryonic CMs with contractile myofibrils showed a change in nuclear organization at day four while non-contractile cells or CMs on stiff substrates do not; scales=5 µm. e) Total RNA was collected after four days of culture. RNAseq analysis revealed that most of the 82 expressed histone genes were downregulated on stiff PDMS. n=4; FPKM: Fragments Per Kilobase of transcript per Million mapped reads. f) Volcano plot of genes associated with the gene ontology term histone methylation (GO:0016571) as determined by RNAseq. Indicated are genes coding for H3K9 methylases, which were amongst the most significantly altered. g) PCR validation of RNAseq (SEQ) data verified downregulation of H3K9 methylating genes and showed that cardiac transcription and structural marker were decreased on stiff substrates. H3K9 demethylase Kdm3a and H3K27-specific methylase Ezh1 showed no change while H3K27 demethylase Kdm6a was downregulated. SD; n=4; T-test (HM=1): * p<0.05, ** p<0.01.
