Fig. 6: Colocalization of H3K9 trimethylated chromatin with actin and emerin is abrogated in CMs on stiff substrates in vitro.
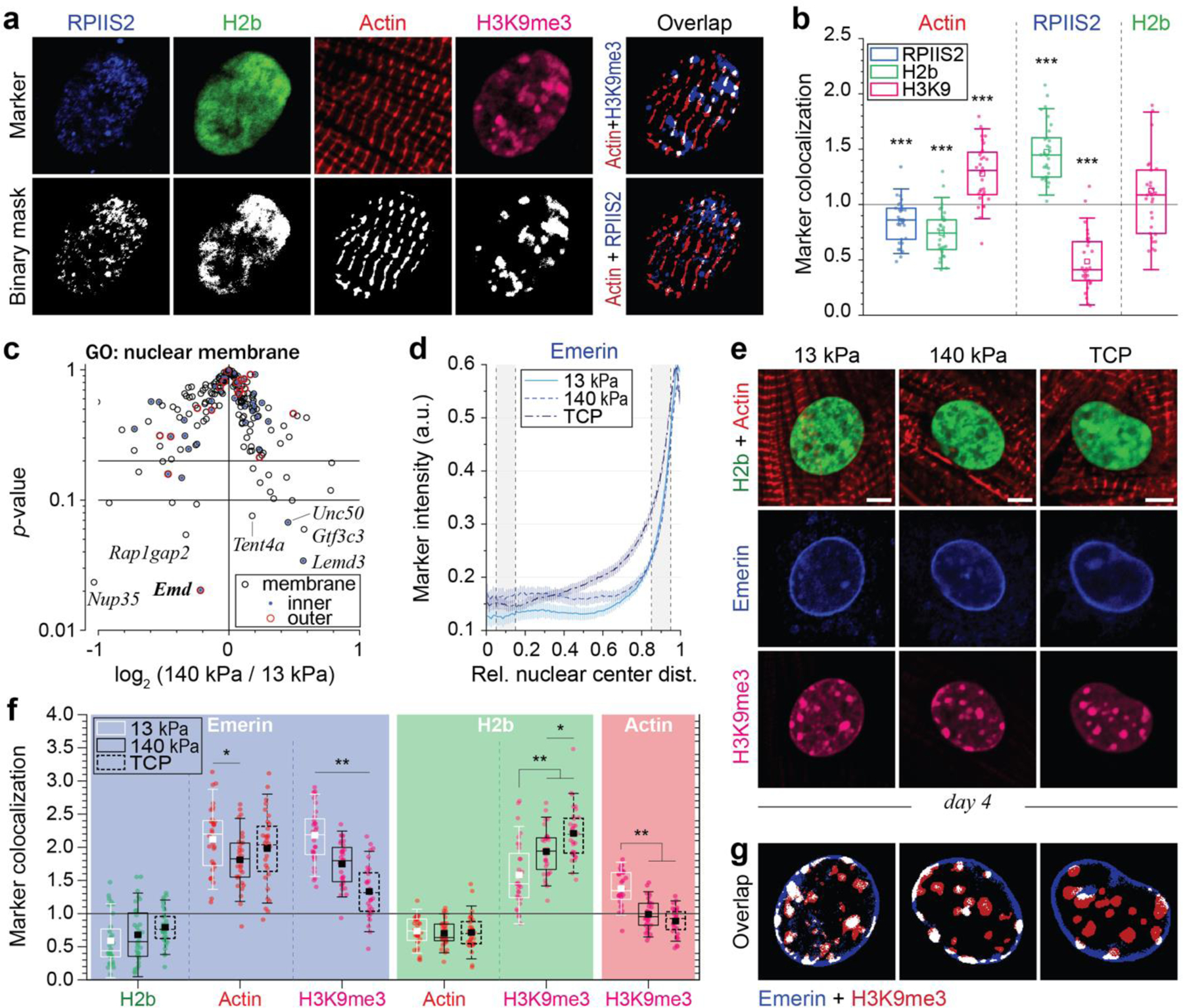
a, b) After four days in culture on soft PDMS, CMs were stained for regions of active transcription (RPIIS2), sarcomeric I-bands (actin) and H3K9me3. Marker channels were binarized and a colocalization score was calculated for each marker pair as the number of overlapping pixels normalized over the number of pixels that would overlap by chance (1=chance). H3K9me3-marked chromatin showed above chance associating with actin while regions of active transcription and overall chromatin did not. n=30 from 3 exp.; T-test (HM=1): *** p<0.001. c) Volcano plot of embryonic CMs, cultured for four days on soft (13 kPa) or stiff (140 kPa) PDMS, showing genes associated with the GO-term nuclear membrane (GO: 0031965), of which Emd (emerin) was the most significantly altered (see also Extended Table 7). n=4. d, e) Embryonic CMs were plated on soft or stiff PDMS or tissue culture plastic (TCP) for four days and stained for emerin, H3K9me3 and actin. Peripheral enrichment analysis showed only minor changes in emerin intensities distribution between all substrates. n=32 from 3 exp. f, g) Colocalization of emerin with H3K9me3 was significantly reduced on stiffer substrates, while no changes in the colocalization with actin was observed. n=32 from 3 exp.; 1W-ANOVA: * p<0.05, ** p<0.01.
