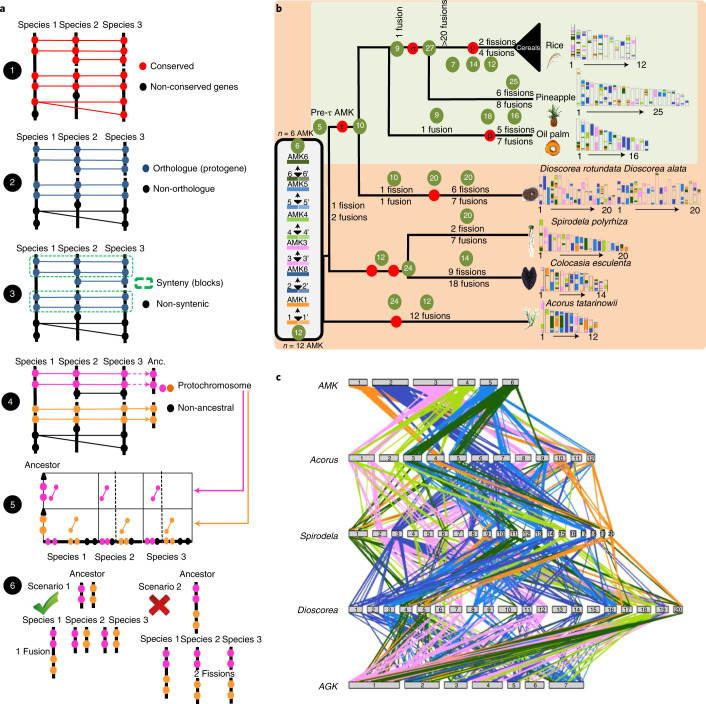
Fig. 4. Monocot genome evolution from the inferred AMK.
a, Illustration of the procedure for reconstructing ancestral karyotypes from conserved genes (step 1), orthologous relationships (step 2), SBs (step 3), CARs (step 4), dotplot validation (step 5) and the best scenario explaining the transition between ancestral and modern genomes (step 6). b, Illustration of the reconstructed AMKs (left), with a six-colour code (with light and dark shades), that evolved into the modern genomes (right) of Acorus, Spirodela polyrhiza, Colocasia esculenta and Dioscorea (alata and rotundata) as well as the AGK (pre-ρ and post-ρ grass ancestors), oil palm (pre-p and post-p ancestors) and pineapple as defined in Murat et al.13 (green panel). Polyploidization events are shown as red dots on the tree branches. The evolution of the number of chromosomes from the AMK to the extant species is shown in green circles together with inferred chromosomal rearrangements (fissions and fusions) on the tree branches. c, Illustration of the synteny between the AMK and modern monocot species with conserved genes linked with lines between chromosomes using the colour code from b.