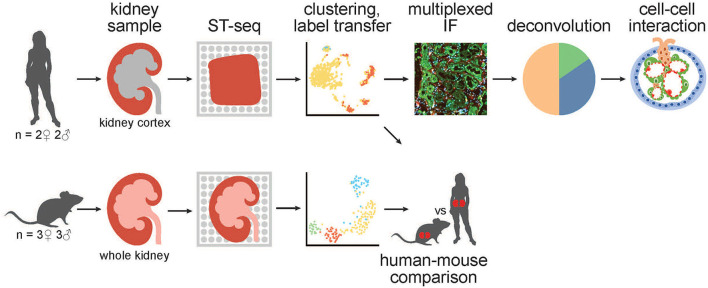
Figure 1.
A schematic of the workflow for generation and analysis of mammalian kidney ST-seq datasets. (From left to right) We performed ST-seq on four human kidney cortical tissues (2 women, 51–53 years old and 2 men, 54 and 56 years old) and six mice whole kidneys (3 males, 8 weeks old and 3 females, 6 weeks old). With the generated ST-seq datasets, we performed clustering and label transfer to define the location of specific nephron segments or regions in human and mouse kidneys, respectively. We selected the cortical kidney regions in the mouse ST-seq datasets and performed functional analysis on genes that were differentially expressed between species in the cortex. In the human cortical kidney tissues, we performed multiplexed IF on consecutive deeper sections to correlate the label transfer annotation of the functional nephron segments with histomorphology. Last, we investigated the CCI by screening for L–R gene pair co-expression in glomerular and PT–PC ST-spots identified by deconvolution.