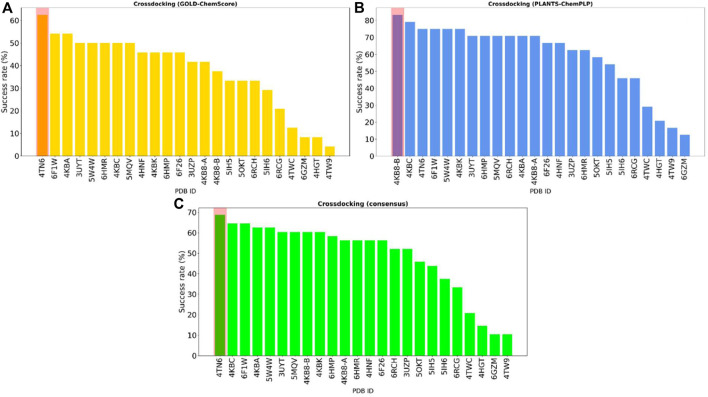
FIGURE 3.
The overall “success rate” in reproducing the correct crystallographic binding mode for each of the 24 CK1δ complexes considered in the study. The “success rate” is defined as the percentage of successful docking runs for each protein in the cross-docking experiment, where a successful docking run is defined as a docking calculation where the RMSD between the best docking pose and the crystal reference falls below an arbitrarily chosen threshold value of 2 Å. Panel (A) reports the results for the GOLD-Chemscore protocol. Panel (B) reports the results for the PLANTS-PLANTSChemPLP protocol. Panel (C) encompasses the combined “success rate” for each protein, defined as the average between the success rate for each protocol. Protein from the complex 4TN6 was chosen as the most representative CK1δ structure for successive calculations.