Fig. 1. T2T-CHM13 assembly supports identification of previously unknown repeat families and complex epigenetic signatures.
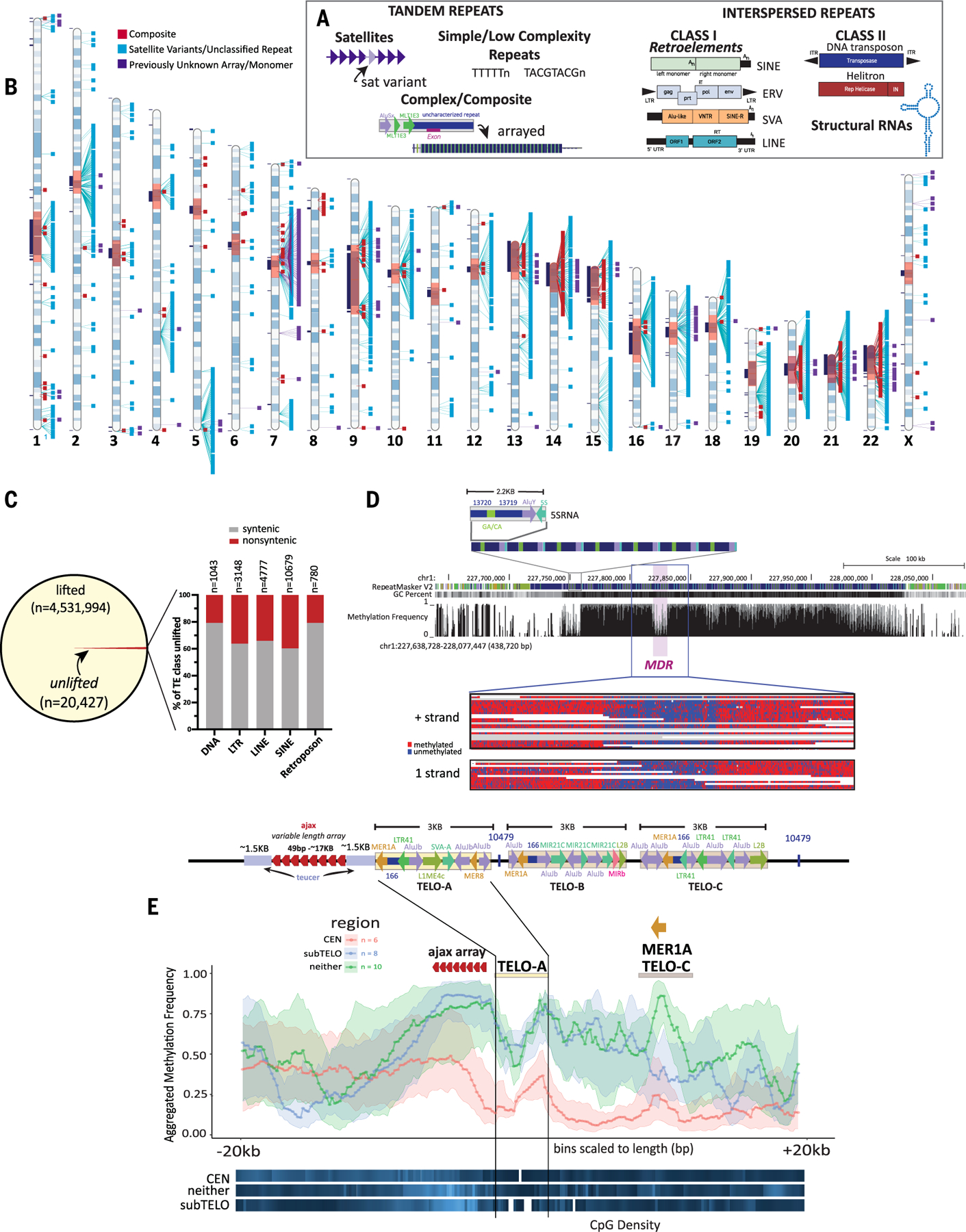
(A) Schematic illustrating examples of tandem repeats, including satellites, simple and low complexity repeats and composites, and interspersed repeats, including class I and class II TEs, and structural RNAs. (B) Ideogram of CHM13 indicating the locations of annotated composite elements (red), satellite variants and unclassified repeats (aqua), and arrays or monomers of sequences found within those arrays (purple). Gaps in GRCh38 with no synteny to T2T-CHM13 (11) are shown in black boxes to the left of each chromosome, centromere blocks [including centromere transition regions (12)] are indicated in orange. (C) (Left) The number of TEs lifted and unlifted from T2T-CHM13 to GRCh38. (Right) Bar plot showing percentage of TEs by class (DNA, LTR, LINE, SINE, and retroposon) that were unlifted from T2T-CHM13 gap-filled regions (nonsyntenic, red) and syntenic regions (gray); the n values show the number of elements within each class affected. (D) (Top) T2T-CHM13 genome browser showing the 5SRNA_Comp subunit structure and array. RepeatMaskerV2 track, CG percentage, and methylation frequency tracks are shown. The MDR is indicated. (Bottom) A zoomed image of individual nanopore reads showing consistent hypomethylation in the MDR (chr1:227,818,289–227,830,789) and hypermethylation in the flanking regions (chr1:227,804,021–227,845,689). Both positive (top) and negative (bottom) strand aligning reads show the same methylation pattern. (E) (Top) Each T2T-CHM13 TELO-composite element consists of a duplication of a teucer repeat (blue) separated by a variable 49-bp (ajax) repeat array (red arrowheads) and three different composite subunits (TELO-A, -B, and -C). Repeat and TE annotations are shown. Some copies of TELO-composite contain the previously unknown repeat “10479” between the TELO-A and TELO-C subunits and/or after the TELO-C subunit. (Bottom) Metaplot of aggregated methylation frequency (average methylation of each bin across the region, 100 bins total) centered on the TELO-A subunit, ±20 kbp, grouped by chromosomal location (orange, centromeric; blue, subtelomeric; green, interstitial). CpG density for each group is indicated at the bottom (white, no CpG; dark blue, low CpG; bright blue, high CpG). The location of the ajax repeat array and the MER1A element within the TELO-C subunit are indicated.
