Fig. 6. Repetitive elements define differences between human genomes and nonhuman primates.
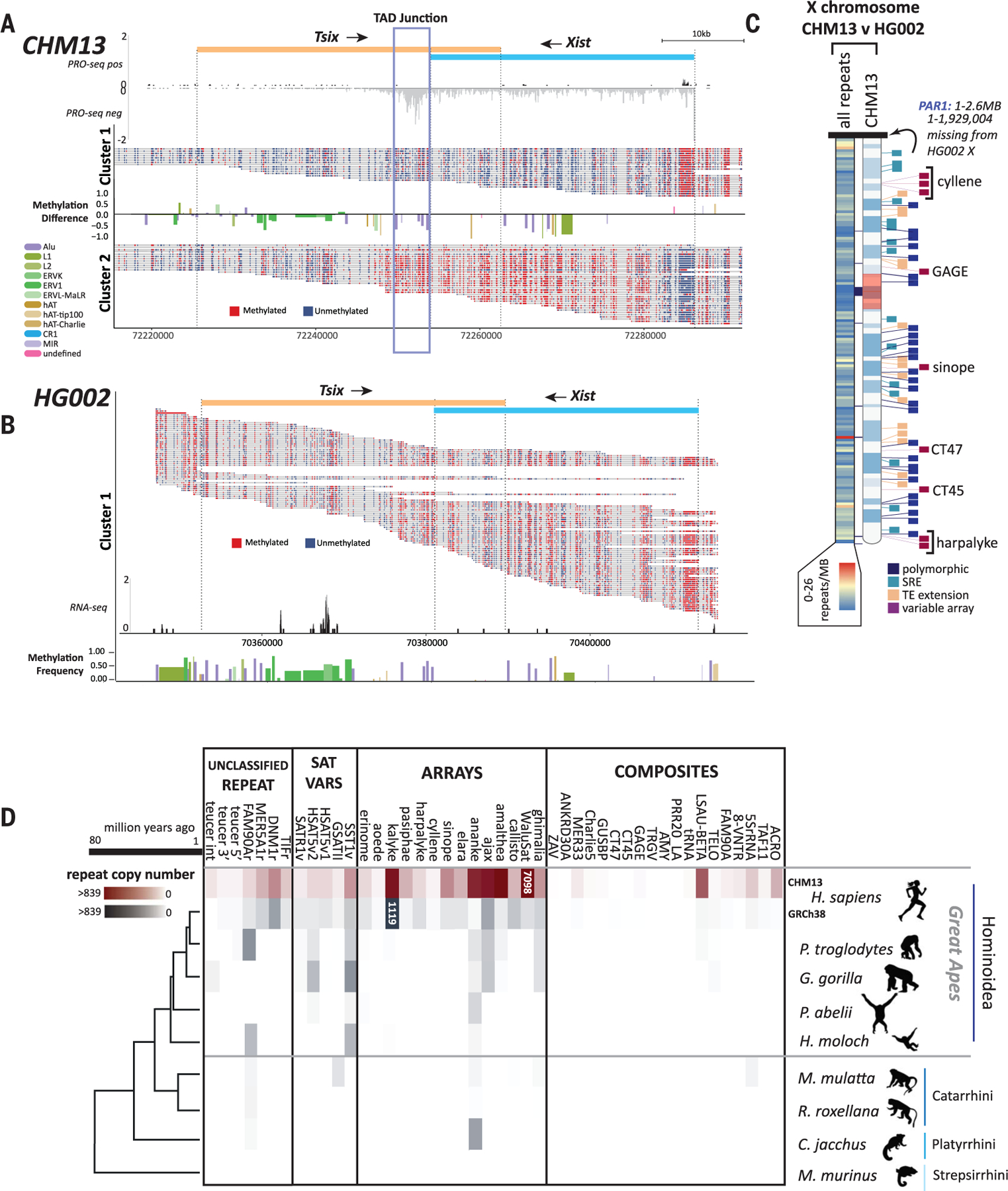
Single read methylation profiles were extracted, and reads were clustered on the basis of the methylation state of the Xist promoter from (A) T2T-CHM13 and (B) HG002. Differences in repeat methylation were calculated by taking the average methylation per repeat and subtracting cluster 2 repeats from cluster 1 repeats. Directionality of Xist/Tsix transcript units are indicated (top). Normalized PRO-seq reads show a marked pileup of RNA pol II at the predicted TAD boundary at the 3′ end of the Xist transcript [(A), blue box]. (B) Normalized RNA-seq reads across the single cluster for HG002 show no transcriptional signal for Xist. (C) Heatmap of chromosome X showing the location of all repeat differences between the Xs of HG002 and T2T-CHM13 (left) and the location of the top four categories of repeat differences: polymorphic (insertion/deletion), SRE (short repeat extension), TE extension, and variable array length (right ideogram). Gaps between T2T-CHM13 and GRCh38 are indicated with black blocks between the heatmap and ideogram. (D) Copy numbers of previously unknown human repeat annotations identified in T2T-CHM13 grouped by repeats, variants of known satellites, tandemly arrayed sequences, and composite element (inclusive of subunits) for T2T-CHM13 (maroon), GRCh38, and genomes for other primates from the Hominoidea, Catarhini, and Platyrrhini lineages (gray). Heatmap scale denotes number of repeats within the array (0 to 839). Array sizes >839 are indicated within colored blocks. Phylogenetic relationship and millions of years since divergence are indicated on the bottom. Not shown: variants of known centromeric satellites [but see (12)] and the repeat annotation for an AluJb (121) fragment, which could not reliably be delineated in copy number from other closely related full-length AluJb elements.
