Fig. 1.
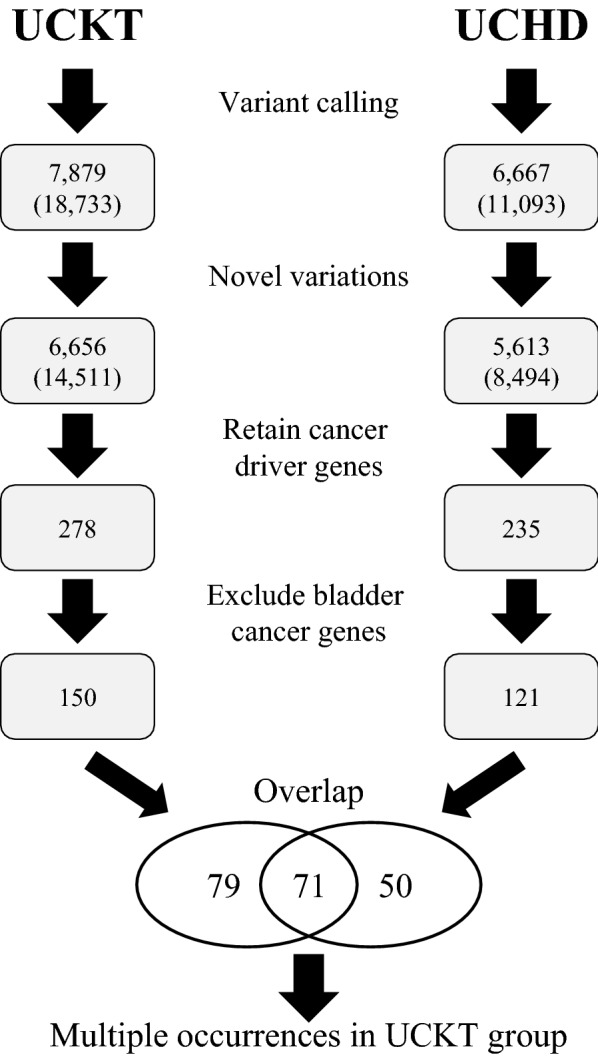
Flow chart of data analysis. The WES data from each patient is subjected to genome mapping and variant calling (Variant calling). Then the data was divided into 2 groups. UCKT; N = 7 and UCHD; N = 5. The mutations were then pooled together in each group and known dbSNPs were excluded (Novel variations). The COSMIC-annotated cancer driver genes were selected (Retain cancer driver genes), but next, bladder cancer-related genes were removed from the list (Exclude bladder cancer genes). Genes that were unique in each group were identified (Overlap). Final list of genes contains those occurred in two or more UCKT patients (Multiple occurrences in UCKT group). Numbers represent gene counts, except those in parentheses, which are nucleotide mutations
