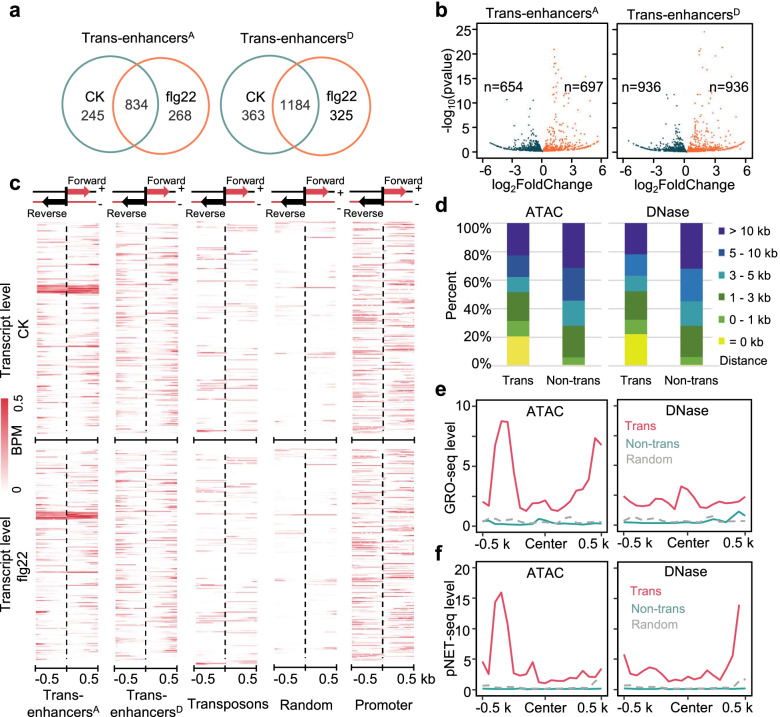
Fig. 1.
Distal enhancer transcription were responsive to flg22 treatment in Arabidopsis. a Numbers of transcribed distal enhancer before and after flg22 signaling activation. Trans-enhancersA, ATAC-seq-based transcribed enhancers; Trans-enhancersD, DNase-seq-based transcribed enhancers. b Volcano Plots show differentially expressed transcribed enhancersA and transcribed enhancersD upon flg22 treatment. The numbers of up (x-axis > 0, warm orange) and down (x-axis < 0, blackish green) transcribed enhancers are shown, respectively. c Heatmaps of transcript levels at transcribed enhancers, transposons, promoters (proximal ATAC-seq peaks), and random sequences. Transcript level with and without flg22 treatment were calculated at and around (±0.5 kb) the sequence candidates. The color scales are in BPM for transcript level. d Distribution of the mean distance from enhancers to the nearest RNA Pol II peaks. e, f Distribution of the signals (BPM scale) derived from GRO-seq (e) and pNET-seq (f) at transcribed enhancers, nontranscribed enhancers, and random sequences. Left and right panels represent ATAC-seq-based enhancers and DNase-seq-based enhancers, respectively