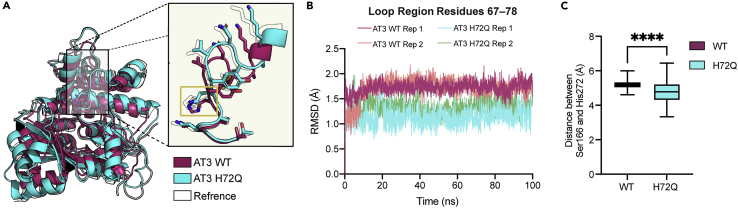
Figure 6.
Molecular dynamic simulations of WT AT3 and AT3 H72Q
(A) Overlaid representative time-averaged structures for AT3 wild type (magenta) and AT3 H72Q (cyan) derived from the last 80 ns of MD simulations. H/Q72 is highlighted with a yellow box. Starting wild-type structure (transparent) shown. Homology models of proteins obtained from AlphaFold2.
(B) RMSD of the Cα from residues 67–78 (loop region) for AT3-E wild type (magenta and peach) and AT3-E H72Q (cyan and green) over 100 ns. Each simulation was performed in duplicate as displayed. Data suggest greater overall residue movements in this region for AT3-E wild type when compared to AT3-E H72Q.
(C) Boxplots showing the distance between residues of the catalytic dyad (Ser166 and His272) measured from the γ-oxygen of S166 to the Δ-nitrogen of His272 from duplicate simulations calculated for the final 80 ns of MD simulations, every 0.1 ns (n = 800). Significance was calculated using an unpaired t-test, ∗∗∗∗p < 0.0001.