Fig. 3. Structural basis for piRNA target binding.
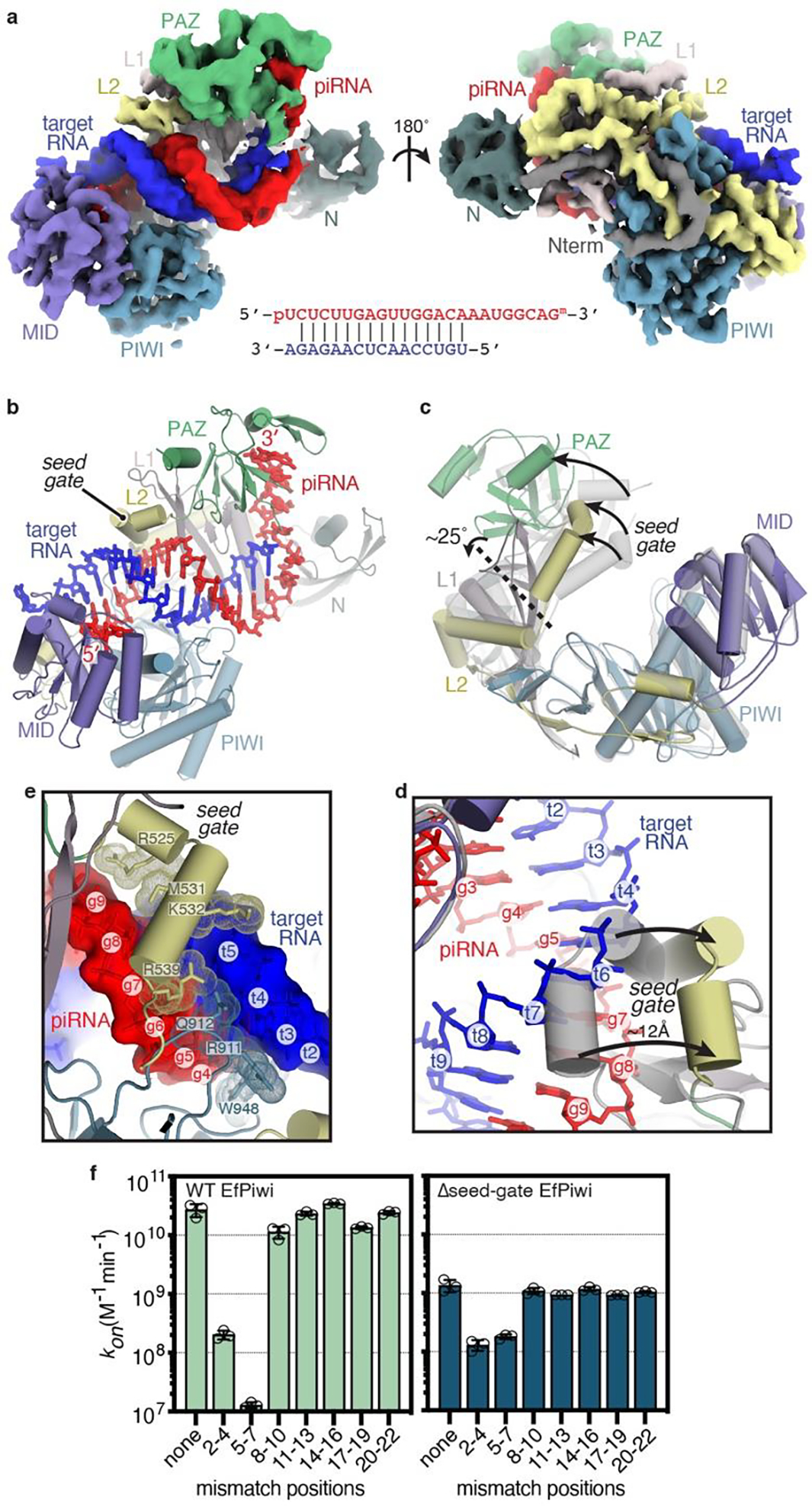
a, Segmented EfPiwi-piRNA-target RNA cryo-EM reconstruction, colored as in Fig 1. piRNA-target base pairing schematic shown below. b, Cartoon representation of EfPiwi-piRNA-target RNA model. c, Superposition of EfPiwi-piRNA (gray, semi-transparent) and EfPiwi-piRNA-target (colored and solid) structures. Arrows indicate movements to target-bound structure. Dashed line indicates hinge in L1 stalk. d, Closeup view showing the seed-gate shifts ~12 Å upon target binding. Arrows indicate direction of movement from guide-only to target-bound conformation. e, Interactions between EfPiwi and the piRNA-target duplex around the seed region. f, Association rates of target RNAs with three consecutive mismatches binding wild-type (left) and Δseed-gate (right) EfPiwi. n=3 independent experiments, data are mean ± s.d.
