Fig. 4. Extensive pairing activates piRNA-target cleavage.
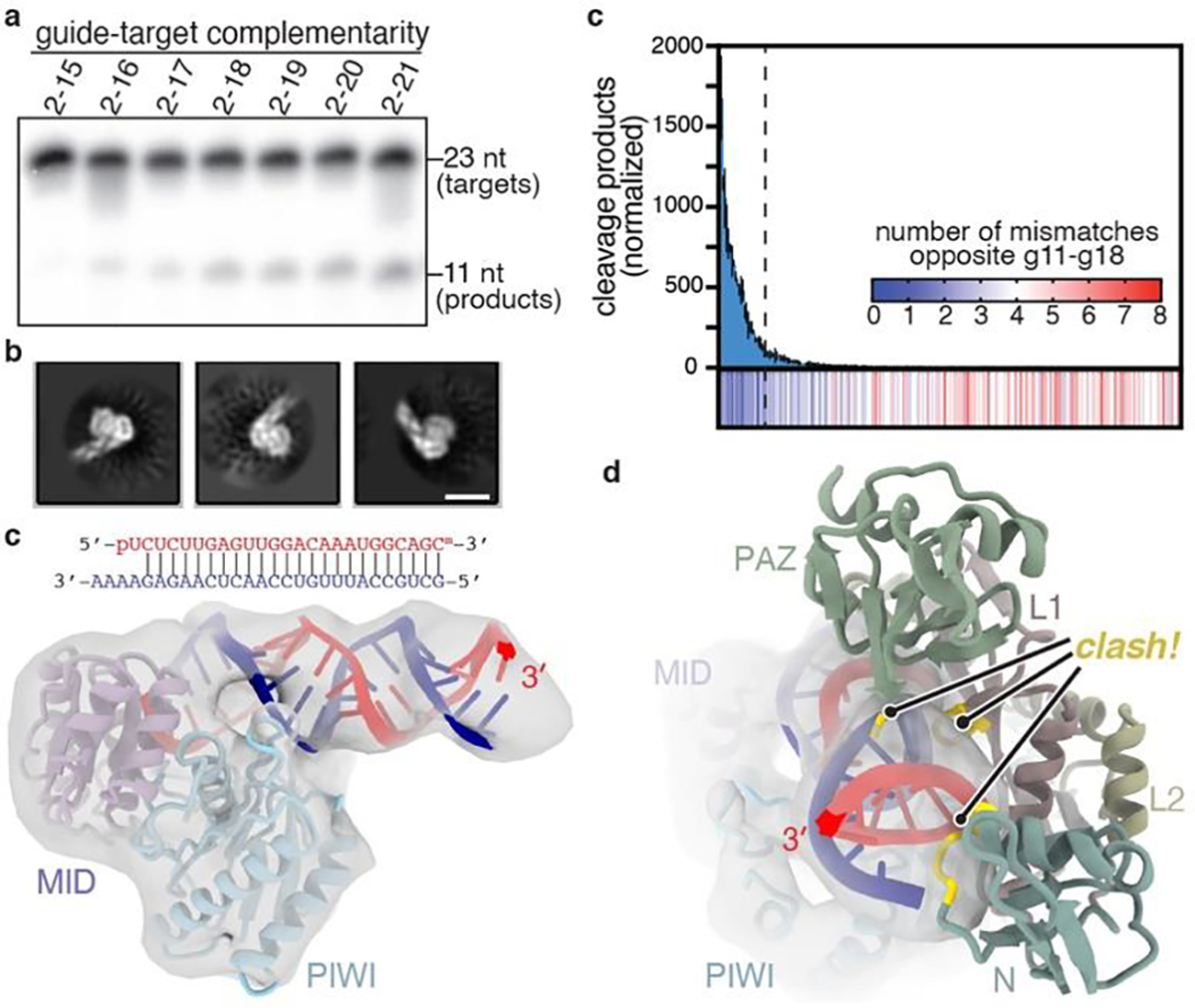
a, Representative denaturing gel showing intact and cleaved target RNAs, with increasing amounts of complementarity to a piRNA, after incubation with excess EfPiwi-piRNA for one hour (results typical of 3 replicates). b, relative number of RNA-seq reads of cleavage products after treating a pool of 256 target RNAs with excess EfPiwi-piRNA. Heatmap indicates number of mismatches opposite piRNA nucleotides g11–g18 in each sequence. Dashed line denotes 26 most abundant products. n=3 independent experiments, data are mean ± s.d. d, Three examples of 2D class averages of EfPiwi-piRNA complex bound to a target with complementarity from g2-g25. Scale bar is 5 nm. e, 3D cryo-EM reconstruction fit with the MID/PIWI lobe and extended piRNA-target duplex. piRNA 3’ end indicated. g, Docking the EfPiwi-piRNA-target model with pairing limited to g2-g16 (see Fig. 3) reveals the N, L1, and PAZ domains sterically clash (gold highlights) with the extended piRNA-target duplex.
