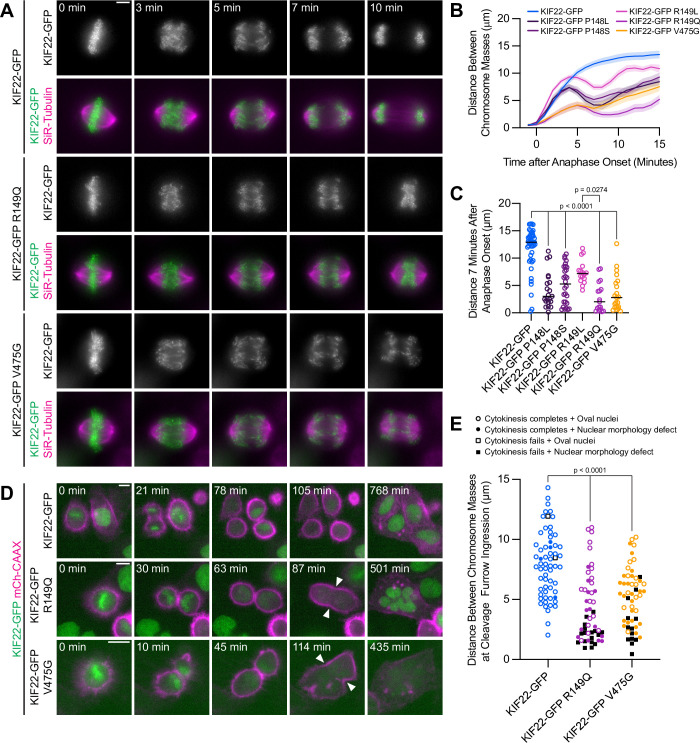
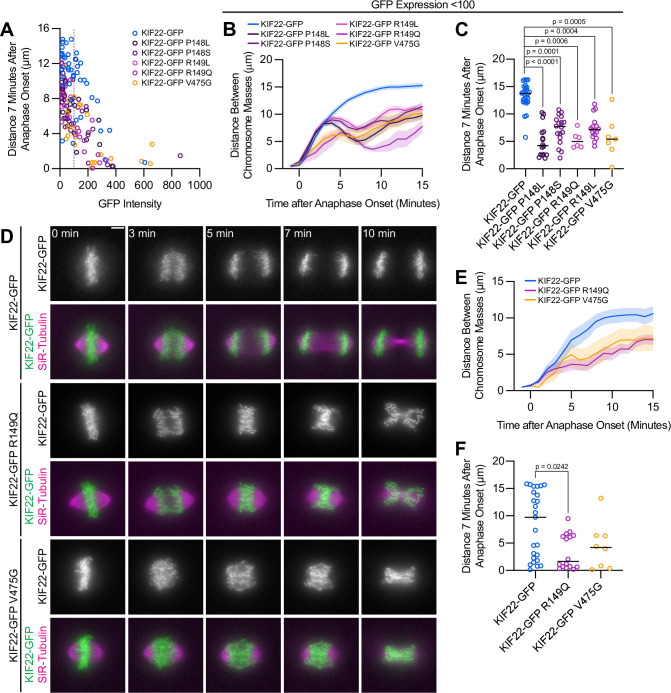
Figure 4. Pathogenic mutations in KIF22 disrupt anaphase chromosome segregation.
(A) Time-lapse images of dividing HeLa-Kyoto cells expressing KIF22-GFP R149Q or KIF22-GFP V475G. Times indicate minutes after anaphase onset. Images are maximum intensity projections in z through the entirety of the spindle. Imaged approximately 18 hr after treatment with doxycycline to induce expression. Scale bar 5 μm. Images are representative of three or more experiments. (B) Distance between separating chromosome masses throughout anaphase in HeLa-Kyoto cells. Lines represent the mean and the shaded area denotes SEM. Forty-three KIF22-GFP cells from 10 experiments, 21 KIF22-GFP P148L cells from 6 experiments, 28 KIF22-GFP P148S cells from 7 experiments, 16 KIF22-GFP R149L cells from 6 experiments, 17 KIF22-GFP R149Q cells from 4 experiments, and 21 KIF22-GFP V475G cells from 21 experiments. (C) Distance between separating chromosome masses 7 min after anaphase onset. Bars indicate medians. P values from Kruskal-Wallis test. P values are greater than 0.05 for comparisons without a marked p value. Data represent the same cell populations presented in (B). (D) Time-lapse images of dividing HeLa-Kyoto cells expressing mCherry (mCh)-CAAX to visualize cell boundaries. Times indicate minutes after anaphase onset. Arrowheads indicate cytokinesis failure. Imaged approximately 8 hr after treatment with doxycycline to induce expression and 24–32 hr after transfection with mCh-CAAX. Scale bars 20 μm. Images are representative of three or more experiments. (E) Distance between chromosome masses at the time of cleavage furrow ingression. P values from Kruskal-Wallis test. P values are greater than 0.05 for comparisons without a marked p value. Sixty-two KIF22-GFP cells from 10 experiments, 52 KIF22-GFP R149Q cells from 9 experiments, and 55 KIF22-GFP V475G cells from 9 experiments. See Figure 4—source data 1.