Abstract
Introduction
C‐peptide is an important marker to assess residual insulin production in individuals with type 1 diabetes (T1D). The accuracy and detection limits of C‐peptide assays are important to detect C‐peptide microsecretion and to reliably observe changes over time in these people. We compared and verified two commercially available assays able to measure C‐peptide in the picomolar range.
Methods
The ultrasensitive Mercodia enzyme‐linked immunosorbent C‐peptide assay (ELISA) was compared with the Beckman immunoradiometric assay (IRMA) for C‐peptide, assessing reproducibility (coefficient of variation [CV]), limit of blank (LoB), limit of detection (LoD) and limit of quantitation (LoQ).
Results
For both assays within‐run and between‐run variation were high at the low (around the detection limit) C‐peptide concentration range, with CVs of around 40%. LoB values for the ultrasensitive ELISA and the IRMA were 1.3 and 0.16 pmol/L respectively. LoD values were 2.4 and 0.54 pmol/L respectively. LoQ values were 9.7 and 3.8 pmol/L respectively. Only the IRMA met the specifications claimed by the manufacturer.
Conclusions
The IRMA provided the lowest threshold for quantification of serum C‐peptide. LoQ of commercially available assays should be established in‐house before applying them in research studies and clinical trials in which low C‐peptide levels have clinical or scientific relevance.
Keywords: biological assay, C‐peptide, diabetes mellitus, limit of detection
Novelty statement.
What is already known?
Low levels of C‐peptide have been found to be associated with better T1D outcomes.
What this study has found?
The performance of two sensitive commercially available C‐peptide assays was verified. One of the assays exceeded the manufacturer's threshold of detection whereas the other did not meet the manufacturer‐specified threshold.
What are the implications of the study?
In‐house verification of manufacturer‐specified performance of available C‐peptide tests for the analysis of low C‐peptide levels and C‐peptide assay standardization is important as C‐peptide is becoming an increasingly important parameter for classification/identification of suitable participants in intervention and prevention studies.
1. INTRODUCTION
C‐peptide has become an increasingly important parameter as it is commonly used as an inclusion criterion for clinical diabetes trials. C‐peptide and insulin pro‐hormone measurements can contribute to sub‐staging T1D (stage 2 [preclinical T1D], stage 3 [new onset T1D] and stage 4 [existing T1D]) and may well become a prerequisite for classification/identification of suitable participants in intervention and prevention studies. 1 Persisting C‐peptide production has been shown to associate with better outcomes and fewer complications, 2 , 3 , 4 , 5 including studies in the Joslin Medalist cohort comprising people with T1D duration >50 years. 6 Very low levels of C‐peptide (~10 pmol/L) were associated with better outcomes. 2 Furthermore, glucagon responses and endogenous glucose production are more pronounced in C‐peptide positive (>50 pmol/L fasting C‐peptide) than in C‐peptide negative (<17 pmol/L) people, suggesting that even low residual β‐cell function may play a role in hypoglycaemia counter regulation. 7
To measure serum C‐peptide a wide range of assays are currently available. We extensively searched the literature for publications reporting C‐peptide measurements and found that in many publications technical validation and verification details of the used C‐peptide assay are often not specified. Moreover, a substantial number of publications does not mention the assay used at all [manuscript submitted for publication]. A possible explanation for this may be that in studies on recent onset diabetes type 1, or in type 2 diabetes, relatively higher residual C‐peptide production will be present than in studies on longstanding type 1 diabetes, and thus measured C‐peptide levels will be well above the detection limits of the well established commercial assays used in random access analysers in routine laboratories. 8 Assay performance should therefore be properly verified and details of this verification should also be reported in publications for correct interpretation of the results. 9
The Mercodia ultrasensitive C‐peptide ELISA is regarded as a reliable test for low‐level detection of C‐peptide by leading T1D research groups and has been reported in many publications, 2 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 but few publications report use of the assay for measuring values near the detection limit. 2 , 11 , 13 , 19 In our clinical laboratory we routinely use a commercially available immunoradiometric assay (IRMA) by Beckman for C‐peptide measurements. According to the manufacturers’ specifications, the Mercodia ultrasensitive C‐peptide ELISA has a lower threshold of detection than the Beckman IRMA. In addition, ELISA methodology is non‐radioactive, less laborious and easy to automate. Hence we considered switching to the Mercodia ultrasensitive C‐peptide ELISA. The main objective of this study is therefore to verify the technical specifications of the ultrasensitive Mercodia ELISA and compare performance with the Beckman IRMA already in routine use.
2. METHODS
The ultrasensitive C‐peptide ELISA assay (Mercodia, cat. no. 10‐1141‐01, Uppsala, Sweden) was compared with the C‐peptide IRMA (Beckman Coulter, cat. no. IM3639, distributed by IMMUNOTECH s.r.o., Prague, Czech Republic). Both assays were performed as described in the manufacturers’ kit inserts. The ELISA plates were read on an Anthos Labtec HTII microtiter plate reader (Anthos labtec) and the IRMA samples were measured on a Wallac Wizard 1470 scintillation counter (Perkin Elmer). Within‐run and between‐run precision were determined by running four pools of predefined target concentrations of approximately 2, 10, 20 and 200 pmol/L, in quadruplicate on five consecutive days, each concentration comprising four samples (i.e. from four individuals) selected based on initial measurements by IRMA.
Samples used in this study were derived from the ‘Biomarkers of heterogeneity in type 1 diabetes’ project (ClinicalTrials.gov Identifier: NCT04977635). The project was approved by the Medical Ethics Review Board of the UMCG. All participants gave written informed consent. People with T1D of 16 years and older with a disease duration of >5 years were included between 2016 and 2019.
The limit of blank (LoB), limit of detection (LoD) and limit of quantitation (LoQ) were determined. 8 The LoB is the highest apparent analyte concentration of a repeatedly measured blank sample (i.e. devoid of the analyte), the LoD is the lowest concentration of the analyte that can be reliably distinguished from the LoB. The LoQ is the lowest concentration at which an acceptable coefficient of variation (CV) is accomplished over an extended period of measurements, ‘acceptable’ meaning within predefined limits of bias and imprecision.
The LoB and LoD were calculated as defined in the Clinical and Laboratory Standards Institute (CLSI) EP17 guideline, 20 using the raw analytical signals (Mercodia: optical density; Beckman: counts per minute). To determine the LoB, we measured two samples devoid of C‐peptide (blank samples: a steroid‐depleted serum sample and serum of a participant with pancreatic agenesis caused by a genetic defect [GATA6 mutation]) 20‐fold in one run. We used the following formula to calculate the LoB:
The LoD is calculated using the LoB and the standard deviation of 20 repeated measures of a low‐concentration sample of approximately 2 pmol/L. The following formula was used:
For the low‐concentration sample a participant sample with a pre‐defined C‐peptide concentration of 2.6 pmol/L, as measured with the IRMA was selected.
After measuring pooled samples with C‐peptide concentrations of 2, 5, 10, 20 and 40 pmol/L in duplicate on five consecutive days, the LoQ was calculated with the LoQ module in EP Evaluator 12 (Data Innovations LLC). For the ultrasensitive ELISA and for the IRMA, curve fits were only possible from a CV of 23% and 20% respectively. A generally accepted CV threshold in the literature is approximately 20%. 21 Intra‐individual CV for C‐peptide is approximately 30%. 22 The World Health Organization (WHO) international standard (ID) 13/146 sample 23 was used as a reference sample and included in duplicate on all plates, which were run in six sessions. The calculated LoB, LoD, LoQ and CV were compared with the limits described in the kit inserts, passing verification when values were equal or lower than the values specified in the kit insert.
3. RESULTS
3.1. Reproducibility
Table 1 shows the within‐run and between‐run precision results. Both assays showed high CV values for the low concentrations (pool 1: target concentration 2 pmol/L; pool 2: target concentration 10 pmol/L). For both assays the within‐run CV values were ≤10% in pool 3 (target concentration 20 pmol/L) and pool 4 (target concentration 200 pmol/L). The between‐run CV values were below 10% in pool 3 and 4 for the IRMA and in pool 4 for the ELISA respectively. The kit insert of neither assay provides reproducibility information in the lower measuring range. For the ultrasensitive ELISA, CV values of concentrations <15 pmol/L are not provided. The lowest concentration for which the IRMA kit insert provides CV values is even much higher at 310 pmol/L (intra‐run) and 290 pmol/L (between‐run). Both the ELISA and IRMA kits show reproducible results (i.e. CV values <20% 21 ) for C‐peptide concentrations of 6.5 pmol/L and above. Of both ELISA and IRMA kits, we also assessed three kit control samples for reproducibility (15, 42 and 111 pmol/L, and 310, 693 and 1428 pmol/L respectively). Within‐run CV values were 6.2/4.6/3.9% and 3.1/2.3/2.7% respectively. Between‐run CV values were 5.4/4.2/2.2% and 5.2/3.6/3.6% respectively.
TABLE 1.
Reproducibility results of the ultrasensitive ELISA and IRMA C‐peptide assays measured in four pool samples with ascending C‐peptide concentrations
| Sample a | ELISA | IRMA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (pmol/L) | Within‐run variation | Between‐run variation | Mean (pmol/L) | Within‐run variation | Between‐run variation | |||||
| SD | CV (%) | SD | CV (%) | SD | CV (%) | SD | CV (%) | |||
| Pool 1 (2 pmol/L) | 0.04 | 0.09 | 255 | 0.1 | 298 | 1.7 | 0.71 | 41.7 | 0.74 | 43.5 |
| Pool 2 (10 pmol/L) | 1.8 | 0.71 | 40 | 0.75 | 42.8 | 5.8 | 0.75 | 12.7 | 1.05 | 18 |
| Pool 3 (20 pmol/L) | 6.5 | 0.66 | 10 | 0.97 | 15 | 11 | 0.55 | 5.1 | 0.99 | 9.1 |
| Pool 4 (200 pmol/L) | 123.3 | 1.35 | 1.1 | 2.84 | 2.3 | 155.7 | 2.1 | 1.4 | 6.99 | 4.5 |
Abbreviations: CV, coefficient of variation; ELISA, enzyme‐linked immunosorbent assay; IRMA, immunoradiometric assay; SD, standard deviation.
Pooled samples with C‐peptide concentrations of predefined targets 2, 10, 20 and 200 pmol/L.
3.2. Detection limit (LoB, LoD and LoQ)
Table 2 shows the results of the LoB and LoD calculations. For the ultrasensitive ELISA the measured serum‐depleted and pancreatic agenesis samples resulted in an LoB of 2 and 1.3 pmol/L respectively. For the IRMA the LoB values were 0.76 and 0.16 pmol/L respectively. These could not be compared with the manufacturers’ data as LoB values are not specified in the kit inserts, but LoB values should be lower than LoD values: the kit inserts of the ELISA and IRMA provide LoD values of 2.5 and 3.79 pmol/L respectively.
TABLE 2.
LoB and LoD calculations
| LoB | LoB (pmol/L) a | LoD kit inserts (pmol/L) c |
|---|---|---|
| ELISA: pancreatic agenesis | 1.3 | ≤2.5 |
| ELISA: serum depleted | 2.0 | ≤2.5 |
| IRMA: pancreatic agenesis | 0.16 | ≤3.79 |
| IRMA: serum depleted | 0.76 | ≤3.79 |
| LoD | LoD (pmol/L) b | |
|---|---|---|
| ELISA: pancreatic agenesis | 2.38 | LOD ≤2.5 |
| IRMA: pancreatic agenesis | 0.54 | LOD ≤3.79 |
Abbreviations: ELISA, enzyme‐linked immunosorbent assay; IRMA, Immunoradiometric assay; LoB, limit of blank; LoD, limit of detection; SD, standard deviation.
Calculated following the EP17 formula LoB = Meanzero sample + 1.645 (SDzero sample) and converted to pmol/L with the calibration curve.
Calculated following the EP17 formula LoD = LoB + 1.645 (SDlow‐concentration sample) and converted to pmol/L with the calibration curve.
LoB values could not be compared to the manufacturers’ data as they are not specified in the kit inserts, but should be lower than the LoD.
The LoD was calculated using the pancreatic agenesis blank sample. With the ELISA assay we measured a lower optical density (OD) for the low‐concentration sample (mean: 0.015 OD) compared with the blank sample (mean: 0.025 OD). This implies high assay variation and poor noise‐to‐signal ratio of the ELISA assay, and thus the robustness of the calculated LoD of 2.38 pmol/L for this assay is highly questionable. For the IRMA assay, higher counts per minute (CPM) were measured with the low‐concentration sample (mean: 65.97 CPM) compared with the blank sample (mean: 43.5 CPM). An LoD of 0.54 pmol/L was calculated, which is well below the LoD mentioned in the manual (3.79 pmol/L).
Figure 1 shows the curve fits to estimate the relationship between mean and CV. Based on the fitted model, the LoQ values for the ultrasensitive ELISA and the IRMA are 9.7 and 3.8 pmol/L respectively. Based on these results, the ultrasensitive ELISA assay did not pass the verification whereas the IRMA did.
FIGURE 1.
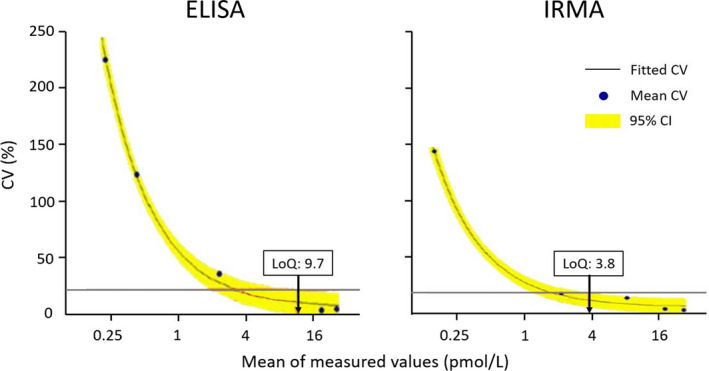
Curve fits by EPE‐12 module, providing LoQ values of 9.7 pmol/L for the ELISA C‐peptide assay with a predefined target‐CV of 23% (red line ELISA) and 3.8 pmol/L for the IRMA C‐peptide assay with a predefined target‐CV of 20% (red line IRMA). CI, confidence interval; CV, coefficient of variation; ELISA, enzyme‐linked immunosorbent assay; IRMA, immunoradiometric assay. Graphs were generated by EP Evaluator 12 (Data Innovations LLC)
The WHO international standard (ID) 13/146 sample was run on all plates of six measurement series. Both assays measured the correct concentration and showed acceptable CV values (Table S1).
4. DISCUSSION
The aim of this study was to assess if the Mercodia ultrasensitive ELISA has a lower LoQ than the Beckman IRMA, as specified by the manufacturers. In the verification of these assays we could not reproduce the manufacturer‐specified LoD of the Mercodia Ultrasensitive C‐peptide ELISA assay. Reproducibility (CV values) were not presented in the kit inserts for concentrations near the detection limit. We found lower CV values for the IRMA when compared with the ELISA. Indeed, the Beckman C‐peptide IRMA used routinely in our laboratory passed the verification, exceeding the manufacturer‐specified LoQ (‘functional sensitivity’ in kit insert) of 13.9 pmol/L. These results are consistent with Oram et al. who also reported that the Mercodia ELISA measured lower values compared with the Roche C‐peptide. 24 Also, the Roche assay could measure C‐peptide in samples in which the Mercodia ELISA could not, despite a lower reported LoD of this assay. Based on our results, we decided to not switch to the ELISA assay and continue with the IRMA for both routine and research purposes, applying an LoQ of 3.8 pmol/L.
A major advantage of the IRMA methodology is the wide measurement range of detection from 3.8 to 6,400 pmol/L (Beckman, cat. no. IM3639 kit insert). Also, radioactive assays are more sensitive than ELISAs while being relatively unaffected by changes made to the chemical composition of samples. 25 The kit insert of the Mercodia Ultrasensitive C‐peptide ELISA specifies neither the measurement range nor the highest calibrator concentration. The latter needs to be acquired from the vial, suggesting it is batch dependent. However, judging from the calibrator curve in the kit insert it seems to be approximately 300 pmol/L. For samples, falling outside the calibration curve of the ultrasensitive ELISA, Mercodia has another, regular C‐peptide ELISA with a lowest calibrator of 100 pmol/L and highest calibrator of approximately 4000 pmol/L (cat. No. 10‐1136‐01). In this case, samples would have to be analysed in two separate tests, posing major disadvantages like use of more volume and repetitive freeze–thaw cycles, offsetting the earlier mentioned advantages of the ELISA methodology.
Overall, our results stress the importance of in‐house verification of technical specifications reported in kit inserts of commercial assays, especially when measuring low concentrations near detection limits. Already in 2008, Little et al. found that C‐peptide measurements acquired by various methods and laboratories do not always agree. 26 Many publications on clinical studies reporting data on C‐peptide measurements do not provide full technical specifications of assay performance (e.g. acceptable CV values are mentioned only for concentrations higher than those measured in the study)[manuscript submitted]. Both LoQ and LoD were specified in the IRMA kit insert, while for the ELISA only the LoD was mentioned by the manufacturer. Moreover, different definitions are used that add to confusion, for example, ‘analytical sensitivity’ instead of LoD, as was the case for the IRMA. However, ‘analytical sensitivity’ is defined as the slope of the calibration curve and is not the same as the LoD. 8 Finally, as the analysers used are also a source of variation, 8 kit inserts should also mention the platform used to determine LoB, LoD and LoQ.
5. CONCLUSIONS
Although even low concentrations of C‐peptide have been shown to correlate with better outcomes, measuring near the detection limit in the context of intervention studies may not be relevant any time soon. However, for mechanistic studies there is an increasing need for being able to measure at or near the detection limits of C‐peptide assays. We compared the Mercodia ultrasensitive ELISA with the Beckman IRMA and found the Beckman IRMA to have superior analytical performance at low C‐peptide concentrations, in contrast to the manufacturers’ details. Our results demonstrate the importance of in‐house verification of manufacturer‐specified performance of laboratory assays, especially when used for a new indication for which clinically meaningful results are outside of the previously used range.
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose. Diabeter is an independent clinic, which was acquired by Medtronic. The research presented here was independently performed and there is no conflict of interest.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank A. Kerdel, laboratory technician at the Department of Clinical Chemistry, IJsselland Hospital, Capelle aan den IJssel, The Netherlands, for his invaluable laboratory work and performing the verification experiments.
de Leur K, Vollenbrock C, Dekker P, et al. How low is really low? Comparison of two C‐peptide assays to establish residual C‐peptide production in type 1 diabetes. Diabet Med. 2022;39:e14785. doi: 10.1111/dme.14785
Funding information
This study was supported by the Juvenile Diabetes Research Foundation (JDRF), grant no. 3‐SRA‐2014‐291‐M‐R, and the Dutch Diabetes Research Foundation, project 2015.16.1856, for which we are very grateful.
REFERENCES
- 1. Rodriguez‐Calvo T, Chen Y‐C, Verchere CB, et al. Altered β‐cell prohormone processing and secretion in type 1 diabetes. Diabetes. 2021;70(5):1038‐1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuhtreiber WM, Washer SLL, Hsu E, et al. Low levels of C‐peptide have clinical significance for established Type 1 diabetes. Diabet Med. 2015;32(10):1346‐1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Luppi P, Drain P. C‐peptide antioxidant adaptive pathways in beta cells and diabetes. J Intern Med. 2017;281(1):7‐24. [DOI] [PubMed] [Google Scholar]
- 4. Jeyam A, Colhoun H, McGurnaghan S, et al. Clinical impact of residual C‐peptide secretion in type 1 diabetes on glycemia and microvascular complications. Diabetes Care. 2021;44(2):390‐398. [DOI] [PubMed] [Google Scholar]
- 5. Rickels MR, Evans‐Molina C, Bahnson HT, et al. High residual C‐peptide likely contributes to glycemic control in type 1 diabetes. J Clin Invest. 2020;130(4):1850‐1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keenan HA, et al. Residual insulin production and pancreatic ss‐cell turnover after 50 years of diabetes: Joslin Medalist Study. Diabetes. 2010;59(11):2846‐2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zenz S, Mader JK, Regittnig W, et al. Impact of C‐peptide status on the response of glucagon and endogenous glucose production to induced hypoglycemia in T1DM. J Clin Endocrinol Metab. 2018;103(4):1408‐1417. [DOI] [PubMed] [Google Scholar]
- 8. Armbruster DA, Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1):S49‐52. [PMC free article] [PubMed] [Google Scholar]
- 9. Leighton E, Sainsbury CA, Jones GC. A practical review of C‐peptide testing in diabetes. Diabetes Ther. 2017;8(3):475‐487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Espes D, Lau J, Carlsson PO. Increased circulating levels of betatrophin in individuals with long‐standing type 1 diabetes. Diabetologia. 2014;57(1):50‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espes D, Singh K, Sandler S, et al. Increased interleukin‐35 levels in patients with type 1 diabetes with remaining C‐peptide. Diabetes Care. 2017;40(8):1090‐1095. [DOI] [PubMed] [Google Scholar]
- 12. Horvaticek M, Djelmis J, Ivanisevic M, et al. Effect of eicosapentaenoic acid and docosahexaenoic acid supplementation on C‐peptide preservation in pregnant women with type‐1 diabetes: randomized placebo controlled clinical trial. Eur J Clin Nutr. 2017;71(8):968‐972. [DOI] [PubMed] [Google Scholar]
- 13. Reinauer C, Rosenbauer J, Bächle C, et al. The clinical course of patients with preschool manifestation of type 1 diabetes is independent of the HLA DR‐DQ genotype. Genes. 2017;8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kalinowska A, Orlińska B, Panasiuk M, et al. Assessment of preservation of beta‐cell function in children with long‐standing type 1 diabetes with "ultrasensitive c‐peptide" method. Pediatr Endocrinol Diabetes Metab. 2017;23(3):130‐138. [DOI] [PubMed] [Google Scholar]
- 15. Steenkamp DW, Cacicedo JM, Sahin‐Efe A, et al. Preserved proinsulin secretion in long‐standing type 1 diabetes. Endocr Pract. 2017;23(12):1387‐1393. [DOI] [PubMed] [Google Scholar]
- 16. Sullivan CA, Cacicedo JM, Rajendran I, et al. Comparison of proinsulin and C‐peptide secretion in healthy versus long‐standing type 1 diabetes mellitus cohorts: a pilot study. PLoS One. 2018;13(11):e0207065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Craig M, Howard N, Silink M, et al. Reduced frequency of HLA DRB1*03‐DQB1*02 in children with type 1 diabetes associated with enterovirus RNA. J Infect Dis. 2003;187(10):1562‐1570. [DOI] [PubMed] [Google Scholar]
- 18. Thivolet C, Marchand L, Chikh K. Inappropriate glucagon and GLP‐1 secretion in individuals with long‐standing type 1 diabetes: effects of residual C‐peptide. Diabetologia. 2019;62(4):593‐597. [DOI] [PubMed] [Google Scholar]
- 19. Wang L, Lovejoy NF, Faustman DL. Persistence of prolonged C‐peptide production in type 1 diabetes as measured with an ultrasensitive C‐peptide assay. Diabetes Care. 2012;35(3):465‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tholen D, Kondratovich M, Armbruster D et al. Protocols for Determination of Limits of Detection and Limits of Quantitation; Approved Guidelines. Oxford Academic; 2004. https://academic.oup.com/clinchem [Google Scholar]
- 21. DeSilva B, Smith W, Weiner R, et al. Recommendations for the bioanalytical method validation of ligand‐binding assays to support pharmacokinetic assessments of macromolecules. Pharm Res. 2003;20(11):1885‐1900. [DOI] [PubMed] [Google Scholar]
- 22. Lacher DA, Hughes JP, Carroll MD. Estimate of biological variation of laboratory analytes based on the third national health and nutrition examination survey. Clin Chem. 2005;51(2):450‐452. [DOI] [PubMed] [Google Scholar]
- 23. Moore M, Dougall T, Ferguson J, et al. Preparation, calibration and evaluation of the First International Standard for human C‐peptide. Clin Chem Lab Med. 2017;55(8):1224‐1233. [DOI] [PubMed] [Google Scholar]
- 24. Oram RA, Jones AG, Besser REJ, et al. The majority of patients with long‐duration type 1 diabetes are insulin microsecretors and have functioning beta cells. Diabetologia. 2014;57(1):187‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cerda‐Kipper AS, Montiel BE, Hosseini S, et al. Immunoassays | Radioimmunoassays and enzyme‐linked immunosorbent assay. In: Worsfold P, ed. Encyclopedia of Analytical Science (Third Edition). Academic Press; 2019:55‐75. [Google Scholar]
- 26. Little RR, Rohlfing CL, Tennill AL, et al. Standardization of C‐peptide measurements. Clin Chem. 2008;54(6):1023‐1026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


