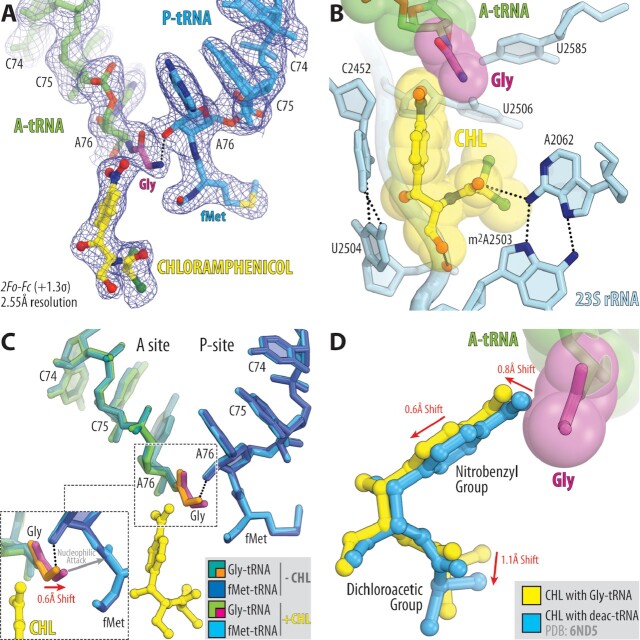
Figure 2.
Structure of the 70S ribosome in complex with CHL and glycyl-tRNA in the A site. (A) 2Fo–Fc electron density map (blue mesh) of the A-site Gly-NH-tRNAGly (green with glycine residue in magenta) and the P-site fMet-NH-tRNAiMet (blue) bound to the T. thermophilus 70S ribosome in the presence of CHL (yellow). (B) Close-up view of CHL bound in the PTC, highlighting H-bond interactions (dashed lines) of the drug with the nucleotides of the 23S rRNA (light blue). P-site tRNA substrate is omitted for clarity. Note that the tip of the nitrobenzyl moiety of CHL forms Van-der-Waals interactions with the glycine residue accommodated in the A site. (C) Superposition of the two new 70S ribosome structures carrying A-site Gly-tRNAGly and P-site fMet-tRNAiMet in the presence and absence of CHL. The structures were aligned based on domain V of the 23S rRNA. Note that binding of CHL causes a 0.6Å shift of the accommodated glycine residue towards the carbonyl carbon of the P-site tRNA substrate (red arrow). (D) Superposition of the previous structure of CHL bound to the 70S ribosome in the presence of deacylated A- and P-site tRNAs (blue, PDB entry 6ND5 (40)) with the new structure of CHL together with the A-site Gly-tRNAGly. Note that, due to the presence of glycine residue, CHL is slightly shifted in its canonical binding pocket to avoid steric hindrance with glycine residue.