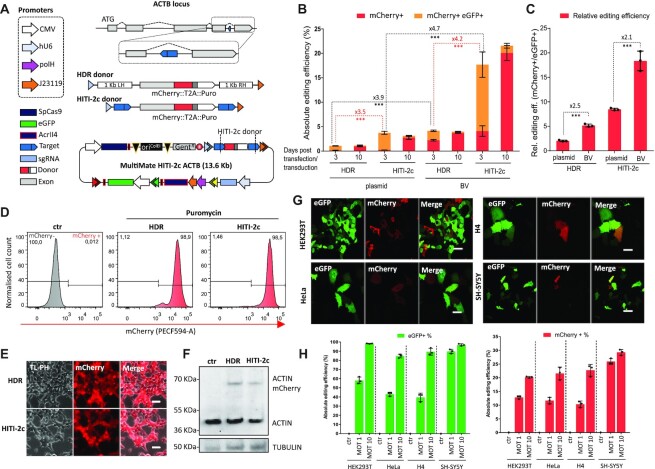
Figure 2.
Baculovirus-vectored delivery of complete multicomponent CRISPR/Cas9 toolkits for homology independent targeted integration (HITI) in human cells. (A) ACTB C-terminal tagging strategy: homologous directed repair (HDR) and homology independent targeted integration (HITI-2c) elements within attL1/attR3 sites (triangles) and MultiMate-HITI-2c ACTB all-in-one DNA circuitry comprising Cas9, HITI-2c donor, sgRNA cassette, eGFP reporter. A module encoding AcrII4 Cas9 inhibitor under control of J23119 and polH promoters ensures vector stability. ACTB C-terminal exon is replaced with a synthetic exon, tagged with mCherry::T2A::puromycin. (B) Absolute gene editing efficiencies of HEK293T cells transfected or transduced with MultiMate-HDR or MultiMate-HITI-2c BVs in the absence of puromycin selection, at three- and ten-days post-transfection/transduction. (C) histogram of relative gene editing efficiencies normalized for transduction/transfection rates at 3 days post transfection/transduction (mCherry + cells %/eGFP + cells %). Histograms represent flow cytometry data. Mean ± s.d. of n = 3 independent biological replicates. ***P < 0.001, Student's t-test. (D–F) HEK293T 21 days after transduction with BV MultiMate HDR or MultiMate HITI-2c BVs after puromycin selection. (D) Representative flow-cytometry histograms. (E) Widefield microscopy, Scalebar = 20 μm. (F) Western blot of total protein extracts. Anti-β-actin antibody was used in top panel with anti-TUBULIN as loading control. (G) Confocal images of HEK293T, HeLa, H4 and SH-SY5Y cells 48 hours after transduction with MultiMate-HITI-2c BV. Scalebar is 50 μm. (H) Histograms of flow-cytometry data of HEK293T, HeLa, H4 and SH-SY5Y 72 hrs after transduction with MultiMate-HITI-2c VSV-G pseudotyped BVs, multiplicity of transduction (MOT) 1 and 10. Transduction efficiency = % of eGFP + cells; absolute gene editing efficiency = % of Cherry + cells. Mean ± s.d. of n = 3 independent biological replicates.