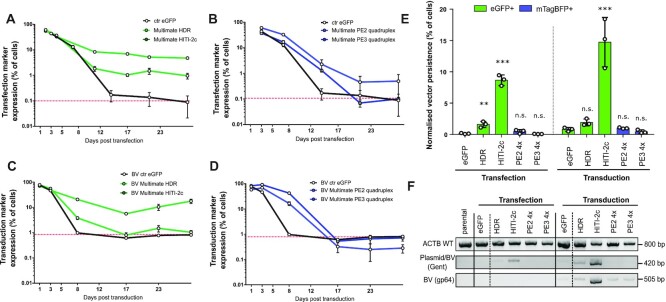
Figure 6.
HDR and HITI, but no prime editing, trigger excess backbone genomic integrations. (A–C) 29 days time-course monitoring of transfection and transduction marker in HEK293T transfected (A, B) or transduced (C,D) with the indicated all-in-one MultiMate constructs harbouring HDR/HITI (A, C) or PE2/3 quadruplex editing toolkits (B, D). A eGFP encoding plasmid or baculovirus, were used as controls for transfections (A, B) and transduction experiments (C, D) respectively. A dashed red line indicates control endpoint detection threshold. Mean with standard deviation of flow-cytometry data of n = 3 independent replicates. (E) Histogram of backbone associated fluorescence markers persistence at 29 days post-transduction normalised by the initial transfection or transduction efficiency. Mean with standard deviations of n = 3 independent replicates. ** P<0.01, ***P < 0.001, n.s.=not significant, Student's t-test. (F) Genomic DNA PCRs of HEK293T at 29 days post-transfection/transduction with the indicated constructs. ACTB amplicon was included to check for DNA quality. Gentamycin (Gent) amplicon detects integration events of both plasmids and baculoviruses, gp64 amplicon detects BV integration events only.