Figure 1.
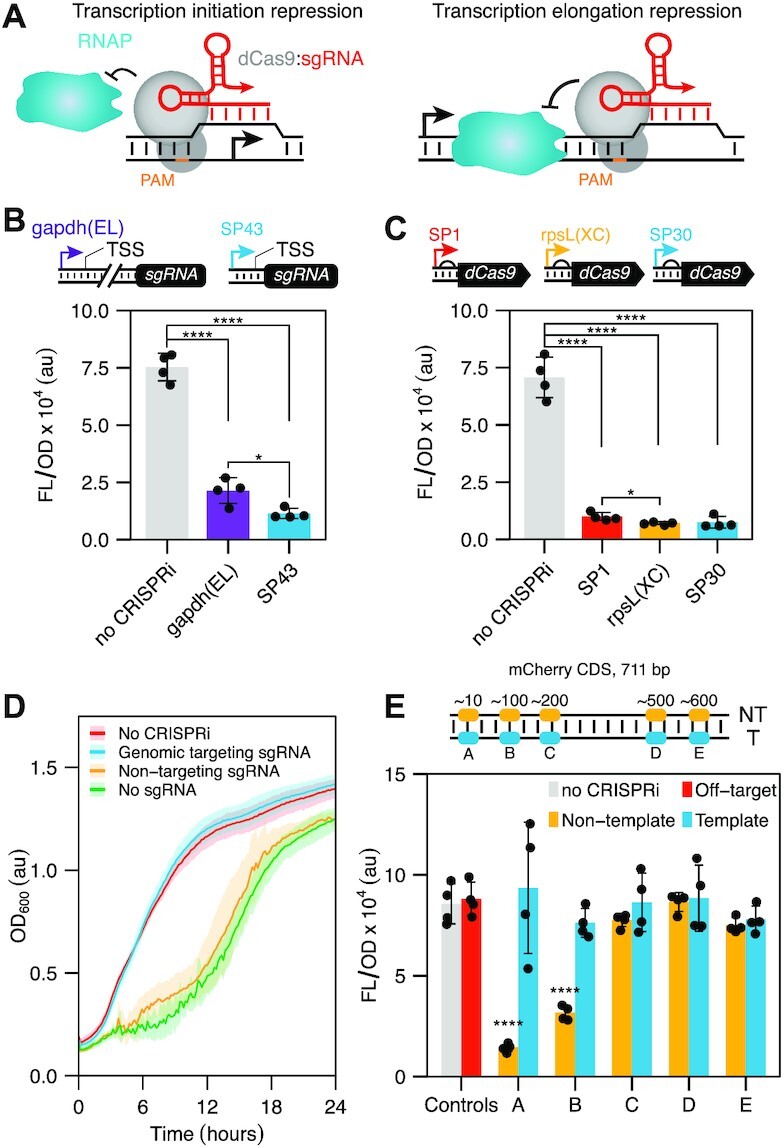
Creating a CRISPR interference (CRISPRi) system for Streptomyces venezuelae. (A) Schematic of CRISPRi mechanism. The ribonucleoprotein complex formed by dCas9 (colored gray) and the single guide RNA (sgRNA, colored red) binds to the promoter or gene coding region to block transcription initiation or elongation by RNA polymerase (RNAP). (B) Eliminating intervening nucleotides between the promoter and the sgRNA leads to higher repression by CRISPRi. Schematic of sgRNA expression cassette with the gapdh(EL) and SP43 promoter. The SP43 promoter contains an annotated TSS, whereas gapdh(EL) TSS remains unannotated. Fluorescence characterization of S. venezuelae cells conjugated with CRISPRi plasmid variants designed to repress transcription of a genomically integrated mCherry reporter. Statistical significance was calculated using two-tailed unpaired Welch's t-test. Statistically significant differences are shown as asterisks (*P-value < 0.05, **** P-value < 0.001). (C) CRISPRi produces robust repression independently of dCas9 expression. Schematic of dCas9 expression cassette using the SP1, rpsL(XC), and SP30 promoters (ascending strength). Fluorescence characterization of S. venezuelae cells conjugated with CRISPRi plasmid variants designed to repress transcription of a genomically integrated mCherry reporter. Statistical significance was calculated using two-tailed unpaired Welch’s t-test. Statistically significant differences are shown as asterisks (*P-value < 0.05, ****P-value < 0.001). (D) Growth rate in the presence of different sgRNAs. Growth was evaluated by measuring optical density (OD) at 600 nm for 24 h. Line shows mean values for each time point. Shaded area shows standard deviation of 4 biological replicates. (E) Position-dependent repression by CRISPRi. Schematic of the sgRNA binding sites used that target different PAMs in the non-template (NT) and template (T) strand of the mCherry reporter gene. Fluorescence characterization of S. venezuelae cells conjugated with CRISPRi plasmid variants. Fluorescence characterization was performed by bulk fluorescence measurements (measured in units of fluorescence [FL]/OD at 600 nm). Data represent mean values and errors bars represent standard deviation of at least three biological replicates. Statistical significance was calculated using two-tailed unpaired Welch’s t-test. Statistically significant differences compared to the no-CRISPRi are shown as asterisks (**** P-value < 0.001).
