Figure 3.
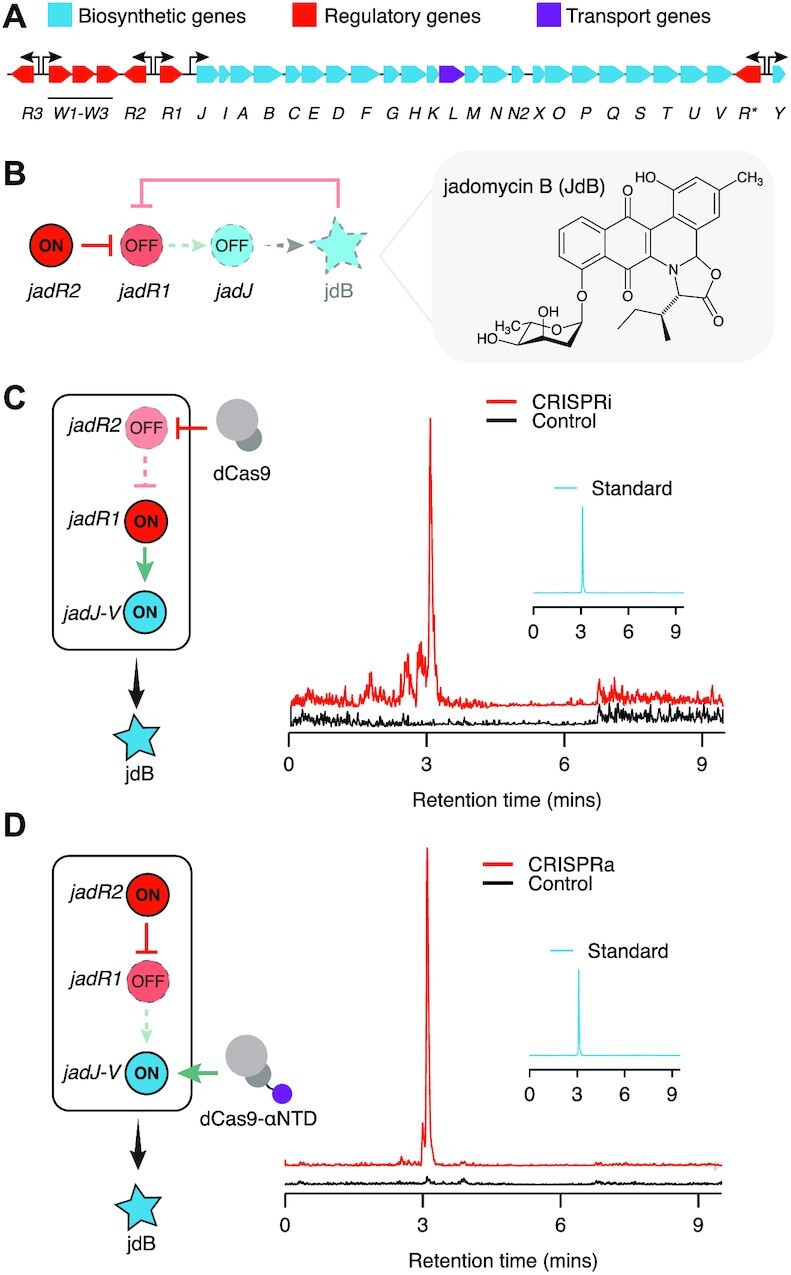
Using CRISPRi and CRISPRa to activate the silent jadomycin b (jdB) biosynthetic gene cluster (BGC). (A) Schematic of the jdB BGC of S. venezuelae. (B) Schematic of JadR1 and JadR2 regulation of the jdB BGC under normal laboratory conditions. Blunted arrows indicate repression and pointed arrows indicate activation. Dotted outline represents an inactive regulator and regulation step. Structure of jdB shown right of panel. (C) Schematic of CRISPRi repressing JadR1 to relieve repression on the jdB BGC and induce expression. LC-MS analysis of extracts from S. venezuelae conjugated with CRISPRi plasmids or no-CRISPRi control. Insert shows LC-MS analysis of a jdB standard. (D) Schematic of CRISPRa activating the jadJ-V operon to induce expression of jdB BGC. LC-MS analysis of extracts from S. venezuelae conjugated with CRISPRa plasmids or no-CRISPRa control. Insert shows LC-MS analysis of a jdB standard. Data in (C and D) are multiple reaction monitoring (MRM) chromatograms at m/z 550.2 → 420.1 ([M + H]+) of one representative biological replicate for each condition. Quantitative analysis is shown in Supplementary Figures S5 and S7. Other biological replicates are shown in Supplementary Figures S6 and S8.
