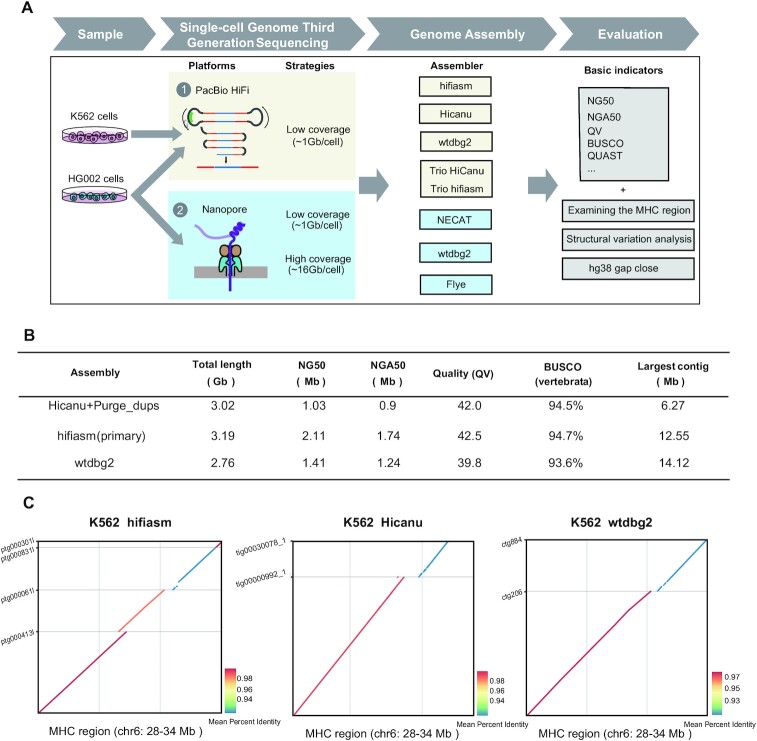
Figure 1.
The assembly workflow and K562 assembly metrics. (A) The workflow illustrates the samples, sequencing platforms, assemblers and evaluation indicators we used to demonstrate the feasibility of genome assembly based on scWGS dataset. (B) K562 assembly (95 cells) benchmarking results for Pacbio HiFi data (primary contigs). N50 is the sequence length of the shortest contig at half of the total assembly size; NG50 is the sequence length of the shortest contig at half of the reference genome size; NGA50 is the sequence length of the shortest aligned block at half of the reference genome size; BUSCO is a tool that assess the completeness of benchmarking universal single-copy orthologs present in an assembly; Per-base consensus quality values (QV) represents a log-scaled probability of errors for assembly, higher QVs indicate more accurate consensus. (C) K562 cells MHC assemblies were compared with the reference human genome (hg38). Only contigs longer than 500kb are displayed.