FIGURE 2.
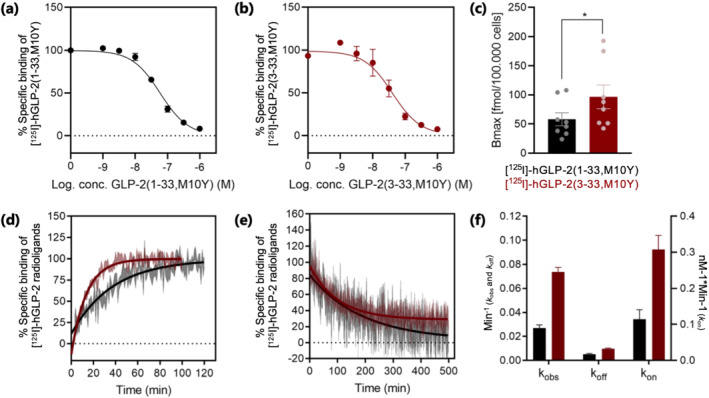
Homologous competition binding and binding kinetic experiments. Homologous competition binding using (a) [125I]‐hGLP‐2(1–33,M10Y) (black) (n = 5) and (b) [125I]‐hGLP‐2(3–33,M10Y) (red) (n = 5). To compensate for inter‐assay variations, data were normalized to the specific binding to hGLP‐2 receptor within each assay. (c) B max for [125I]‐hGLP‐2(1–33,M10Y) (black) and [125I]‐hGLP‐2(3–33,M10Y) (red), (d) association (n = 4) and (e) dissociation (n = 4) of [125I]‐hGLP‐2(1–33,M10Y) (black) and [125I]‐hGLP‐2(3–33,M10Y) (red) on/from the hGLP‐2 receptor. The dissociation was initiated by the addition of 1 μM unlabelled hGLP‐2(1–33,M10Y) or hGLP‐2(3–33,M10Y). (f) Comparison of binding kinetic parameters between [125I]‐hGLP‐2(1–33,M10Y) (black) and [125I]‐hGLP‐2(3–33,M10Y) (red) obtained from association and dissociation assays. The parameters k on, k off, k obs and K D were calculated from the sum of all data, and during this process, the errors were propagated
