Figure 4.
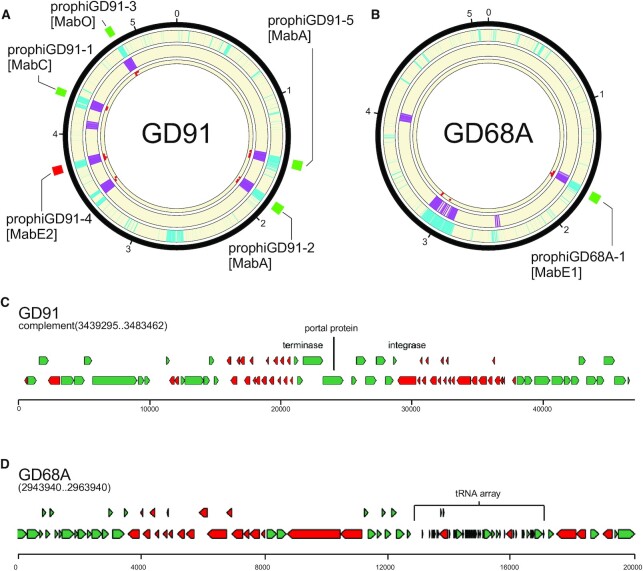
Phage gene homology detection in normal mode eliminates false positives from fast mode. (A) A circular genome diagram for M. abscessus GD91, with five known prophages, illustrated with green (forward oriented) or red (reverse oriented) boxes outside the circle. The outermost track (cyan) shows the location of genes determined not to be part of the M. abscessus shell genome. The next track (purple) shows the location of genes with size/tdc similar to temperate phages. The innermost track (red dots) shows the location of genes with strong hits to one or more HMMs in the limited database used in normal mode. Regions with overlapping cyan and purple signal are identified as prophages in fast mode. Any such regions with fewer than 5 phage gene homologs are removed from the output in normal mode. (B) The same as (A), but for M. abscessus GD68A, which has one known prophage. (C) Close inspection of the false positive from GD91 reveals that it is a defective phage with many of the genes related to Cluster MabI prophages, but 55 kb of the 80 kb MabI length missing, including all tail assembly genes. (D) Inspection of a false positive from GD68A reveals an abundance of short genes arranged in long operons with few tdc's, and an array of 35 predicted tRNA genes that nearly doubles the species normal ∼49 tRNA genes (Supplementary Table S5); nonetheless the region contains no compelling hits to any phage proteins.