Figure 5.
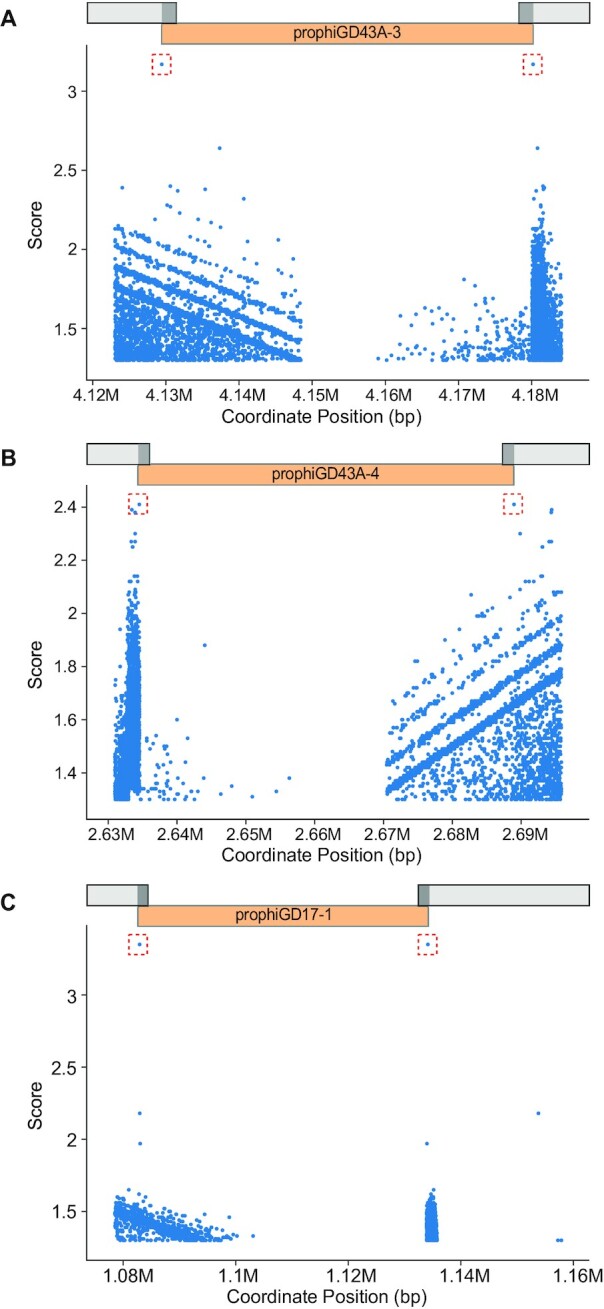
DEPhT identifies attachment sites at prophage boundaries. DEPhT uses BLASTN to identify all sequence repeats present at both the left and right sides of predicted prophages. All left/right pairs identified by BLASTN are scored as described (see Methods) and the highest scoring sites are shown in a red box. The position of the prophage is shown. Three examples are presented: (A) prophage prophiGD43A-3 that uses a tyrosine integrase and has a 17 bp att site common core with a single mismatch between attL and attR, (B) prophage prophiGD43A-4 which uses a serine integrase and has an 8 bp common core at attL and attR, and (C) prophiGD17-1 which uses a tyrosine integrase and has a 39 bp common core without mismatches at attL and attR.
