FIGURE 1.
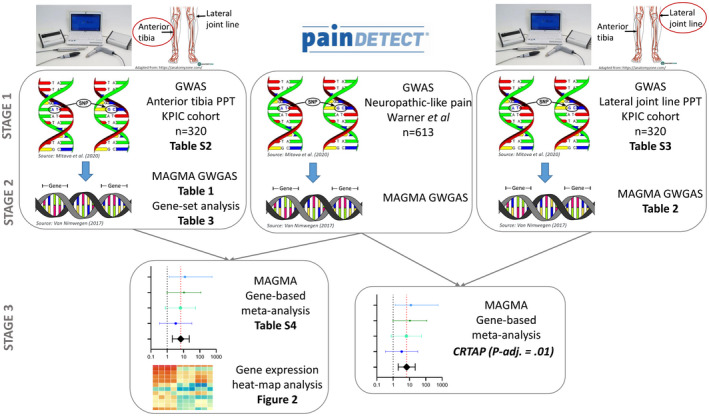
Study design. A three‐stage design for identification of associations between genetic variants and two pain sensitivity phenotypes: distal sensitization [anterior tibia pressure pain threshold (PPT)] and local sensitization (lateral joint line PPT), as well as genes common in pain sensitivity and neuropathic‐like pain (painDETECT questionnaire) phenotypes. First, two GWASs with PPTs from both distal and affected sites were run to identify genetic variants for distal and local sensitization, respectively, using data from the KPIC cohort (Tables S2 and S3). Second, three GWGASs and gene‐set analyses were run to identify the genes associated with the two pain sensitivity phenotypes and a neuropathic pain‐like phenotype using the GWASs outputs from KPIC and existing GWAS data from a Nottingham cohort (Tables 1, 2, 3). Third, two meta‐analyses of our GWGAS findings were conducted to identify genes common in pain sensitization (distal and local) and neuropathic‐like pain (Table S4) and a heat map analysis of the common genes identified from the distal sensitization/neuropathic‐like pain meta‐analysis was performed (Figure 2)
