Figure 7.
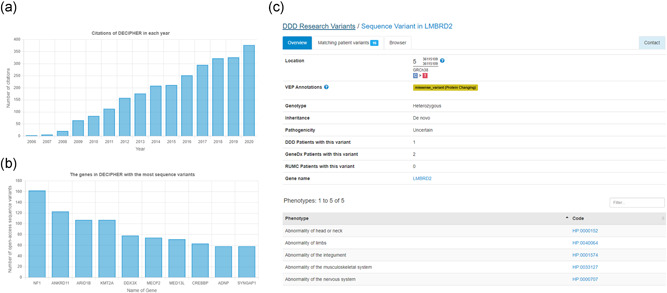
(A) Since its inception in 2004, DECIPHER has been cited in more than 2600 publications. (B) The genes with the most open‐access sequence variants in DECIPHER (at the time of writing). (C) DECIPHER openly shares variants of unknown significance identified in undiagnosed probands in the Deciphering Developmental Disorders study (research variants). For each variant, a page provides details of the variant and high‐level phenotype terms. The number of patients with each variant in the DDD data set is displayed, in addition to the number of patients identified in the GeneDx and Radboud University Medical Center de novo variant data set as described by Kaplanis et al. (2020)
