Figure 1.
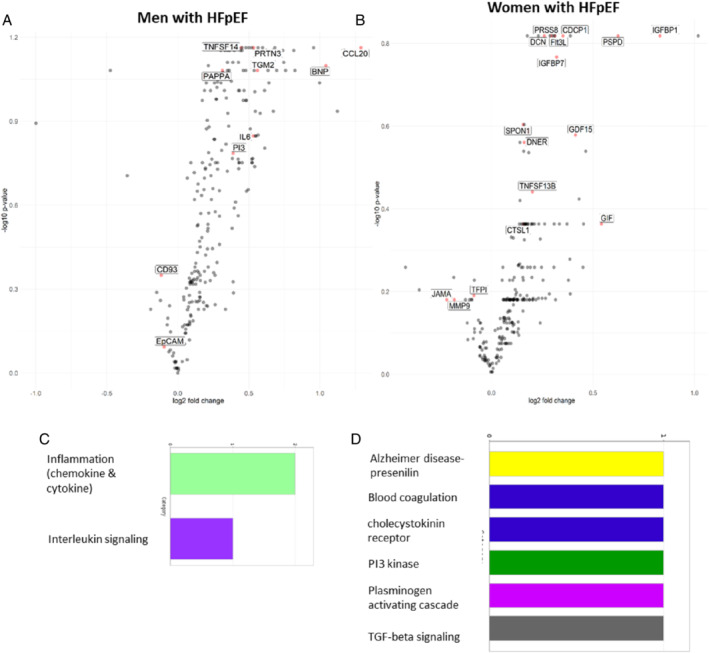
Volcano plots in men (A) and women (B) with heart failure and preserved ejection fraction (HFpEF) showing biomarkers (labelled) selected to be associated with coronary microvascular dysfunction (CMD) (coronary flow reserve [CFR] < 2.5) by lasso penalized regression analyses. The y‐axis shows the −log10 of the false discovery rate‐corrected p‐values for the associations of individual biomarkers with CMD (CFR <2.5) versus no CMD. The x‐axis shows the log2 of the fold changes of the respective biomarker differences between individuals with and without CMD. Bar graphs show functional classification analyses of differentially expressed proteins in men (C) and women (D) with HFpEF. BNP, brain natriuretic peptide; CCL20, chemokine ligand 20; CD93, cluster of differentiation 93; CDCP1, CUB domain‐containing protein 1; CTSL1, cathepsin L1; DCN, decorin; DNER, delta/notch‐like epidermal growth factor‐related receptor; EpCAM, epithelial cell adhesion molecule; Flt3L, FMS‐like tyrosine kinase 3 ligand; GDF15, growth differentiation factor 15; GIF, gastric intrinsic factor; IGFBP1, insulin‐like growth factor‐binding protein 1; IGFBP7, insulin‐like growth factor binding protein 7; IL6, interleukin‐6; JAM‐A, junctional adhesion molecule A; KIM1, kidney injury molecule 1; MMP9, metalloproteinase‐9; PAPP‐A, pregnancy‐associated plasma protein A; PI3, elafin; PRSS8, prostasin; PRTN3, proteinase 3; PSPD, phage shock protein D; SPON1, spondin‐1; TFPI, tissue factor pathway inhibitor; TGM2, transglutaminase 2; TNFSF13B, tumour necrosis factor receptor superfamily member 13B; TNFSF14, tumour necrosis factor receptor superfamily member 14.
