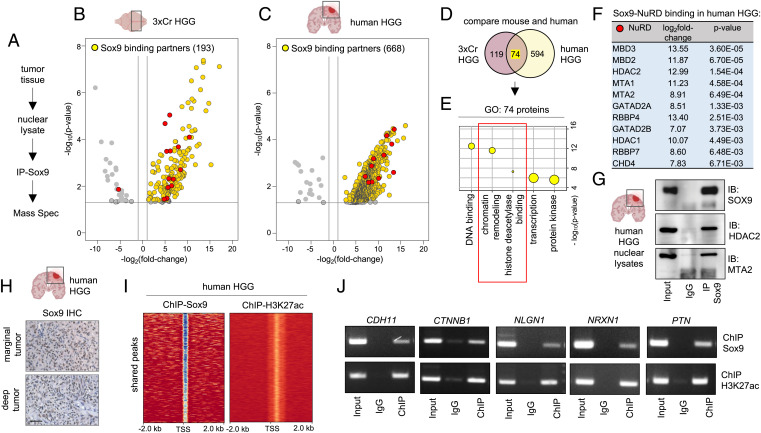
Fig. 4.
Sox9 interacts with histone deacetylation complex in HGG. (A) Schematic of IP-Sox9 and MS proteomic experiment to identify Sox9 binding partners. (B and C) Volcano plots depicting IP-MS data of Sox9 interactome in (B) 3xCr HGG and (C) human HGG. Fold change was calculated over control samples of nuclear lysates incubated with beads only without antibody. IP-MS experiments were performed in independent biological triplicates for 3xCr HGG and two independent human patient HGG samples (P < 0.05 and fold change of >2). (D) Venn diagram showing the number of unique and overlapping Sox9 binding partners in mouse and human HGG. (E) GO terms associated with the 74 shared Sox9 interactors in mouse and human HGG showing enrichment of histone deacetylation. (F) Table showing Sox9 binding fold change and P values with NuRD complex members. Note all NuRD members are depicted by red circles in volcano plots shown in B and C. (G) Sox9 coimmunoprecipitation with NuRD members Hdac2 and Mta2 from human HGG nuclear lysates. (H) IHC of Sox9 in human HGG marginal and deep tumors (brown: Sox9; blue: hematoxylin counterstain; scale bar: 50 μm). (I) ChIP heatmaps at 2 kb from peak center of ChIP experiments with Sox9 and H3K27ac from human HGG tissue, showing that 9,539 of identified H3K27ac peaks are also co-occupied by Sox9 peaks. (J) ChIP-PCR validation of a subset of Sox9 and H3K27ac cotargets in human HGG. ChIP-PCR primer sequences are listed in Dataset S8.