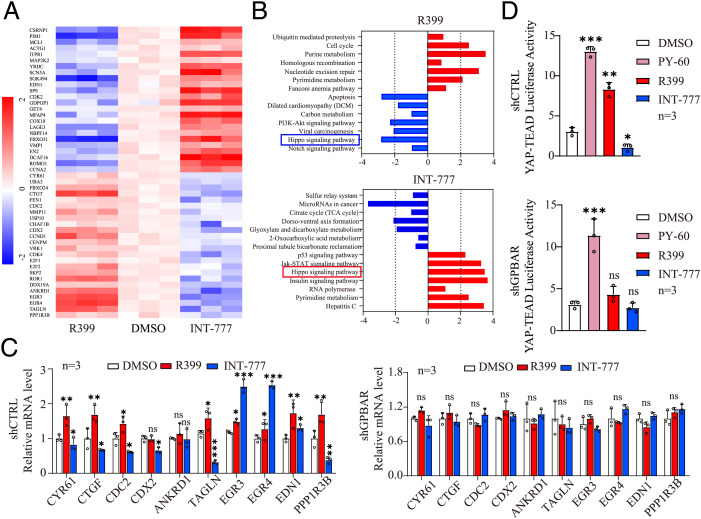
Fig. 3.
Transcriptome analysis reveals that R399 activates but INT-777 inhibits Hippo-YAP signaling in H1299 cells. (A) Heat map of differentially expressed genes (DEGs) in R399-treated and INT-777–treated H1299 cells compared to vehicle-treated cells, respectively. Red indicates up-regulated genes (log2 fold change > 1 and P < 0.05). Blue indicates down-regulated genes (log2 fold change < −1 and P < 0.05). (B) Pathway analysis of DEGs in R399-treated and INT-777–treated H1299 cells. (C) Expression analysis (qRT-PCR) of the indicated YAP target genes in H1299-shGPBAR cells and its control cells stimulated with R399 (700 nM) or INT-777 (2 μM) for 24 h (n = 3). (D) Effects of PY-60 (10 μM), R399 (700 nM), or INT-777 (2 μM) on YAP-TEAD transcriptional activity were determined by the TEAD luciferase reporter assay in H1299-shGPBAR cells and control cells for 24 h (n = 3). Data are presented as the mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 based on the Student’s t-test. CTRL, control; DMSO, dimethyl sulfoxide; mRNA, messenger RNA; ns, not significant; TCA, taurocholic acid.