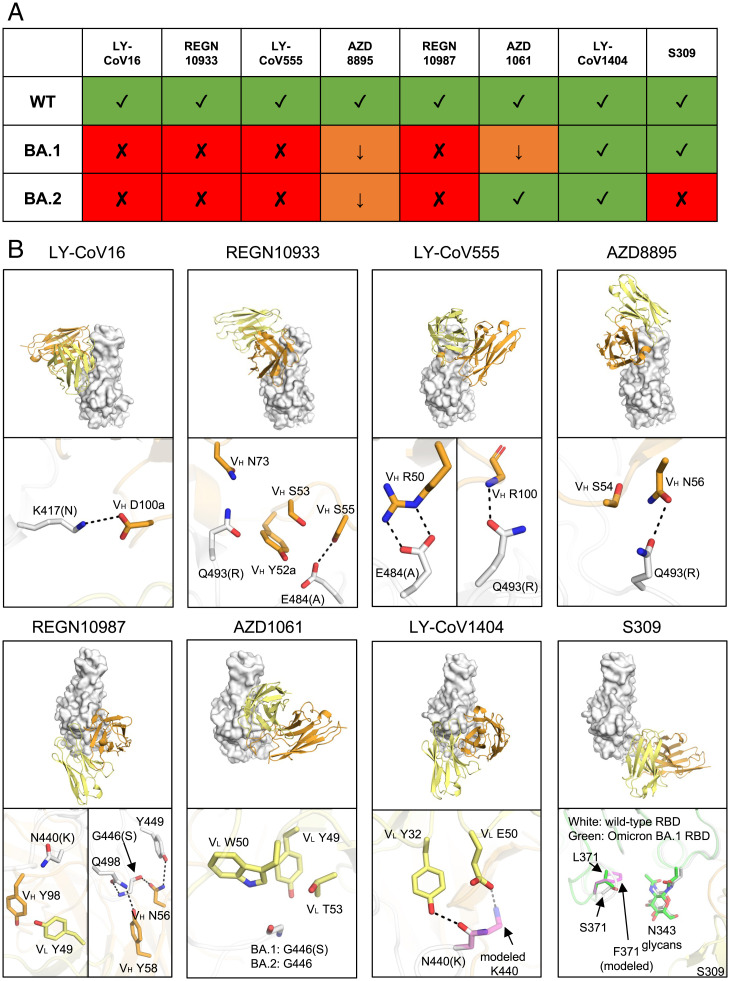
Fig. 4.
Mechanisms of escape or resistance to therapeutic antibodies against SARS-CoV-2. (A) Neutralization activity of the therapeutic antibodies shown are taken from refs. 18, 30, 31. Variants that retain neutralization activity are represented by green “✓” symbols, while variants with reduced or abolished neutralization are indicated by orange “↓” and red “✘” symbols, respectively. (B) Structural explanation of escape mechanisms of the Omicron BA.1 and BA.2 sublineages. RBDs are represented by white surfaces, while the heavy and light chains of the antibodies are in orange and yellow cartoon representation. Hydrogen bonds and salt bridges are shown as black dashed lines. For clarity, only the variable domains of the antibodies are shown. Orientations of the RBDs are the same for all the eight antibodies, as follows: LY-CoV016 (PDB 7C01), REGN10933 (6XDG), LY-CoV555 (7KMG), AZD8895 (7L7D), REGN10987 (6XDG), AZD1061 (7L7E), LY-CoV1404 (7MMO), and S309 (7R6W). The structure of S309 in complex with Omicron BA.1 RBD (green, PDB 7TN0) was superimposed onto the S309/WT-RBD structure for comparison. Mutations S371F for S309-bound RBD as well as N440K for LY-CoV1404-bound RBD are modeled and represented by transparent magenta sticks. Kabat numbering is assigned to all antibody residues.