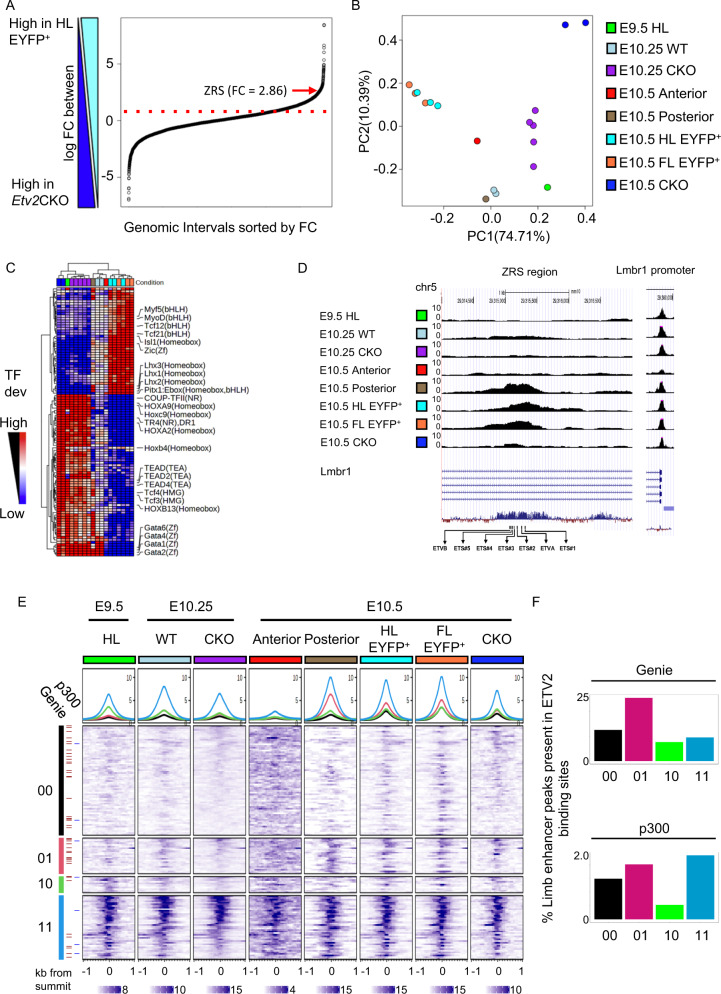
Fig. 3. ETV2 globally affects chromatin accessibility in the limb progenitors, and the ZRS is one of the targets.
A A plot of 5,178 conserved (>0.8) ATAC-seq intervals sorted by the fold change (FC) between HL Etv2-EYFP+ versus Etv2CKO samples. B PCA of different cell populations obtained from limb buds. Each dot represents an ATAC-seq sample (the color labels of the samples are shown in the right panel and are used throughout the figure). PC1 (explained variance: 74.71%) separated the Etv2-expressing (orange and teal dots) and non-expressing populations (blue, purple, and green dots) efficiently (note the separation of dots along the PC1 axis). C The heatmap of transcription factor (TF) deviations showed the variations of TF associated chromatin accessibility in each sample. Each row represents a TF motif, and each column represents an ATAC-seq sample. The row and column ordering were based on the clustering of the TF deviation scores derived by chromVAR. Dendrogram shown at the top reveals the global difference in TF accessibility between Etv2-expressing cells and non-expressing cells. D ATAC-seq analysis of the ZRS region (chr5: 29,314,591 – 29,316,266) (left panel) and the Lmbr1 gene promoter region (chr5:29,377,922-29,379,008) (right panel). Locations of putative ETS and ETV binding sites are indicated at the bottom. E The heatmap shows the ATAC-seq density within 1 kb up- and down-stream of 7,352 putative Etv2 binding sites at different samples. The ETV2 motifs-centric regions were split into four groups according to their chromatin accessibility in E9.5 HL and E10.5 posterior, two samples that does not or does express Etv2, respectively. The regions are split into four groups: closed in both E9.5 HL and E10.5 Posterior (denoted as 00); closed in E9.5 HL and open in E10.5 Posterior (denoted as 01); open in E9.5 and closed in E10.5 Posterior (denoted as 10); and open in both E9.5 HL and E10.5 Posterior (denoted as 11), respectively. F The bar plot shows the proportion of the known limb enhancers from Limb-Enhancer Genie45 and the published p300 ChIP-seq peaks in each cluster, respectively. Each sample in panels B, C, D and E is represented by the same color. For wild-type samples, dissected hindlimb buds from 16 to 20 embryos were pooled for each sample. For Etv2CKO, samples from 3 to 5 embryos were pooled. Sex of the embryos were not determined. Each track represents average of two replicates.