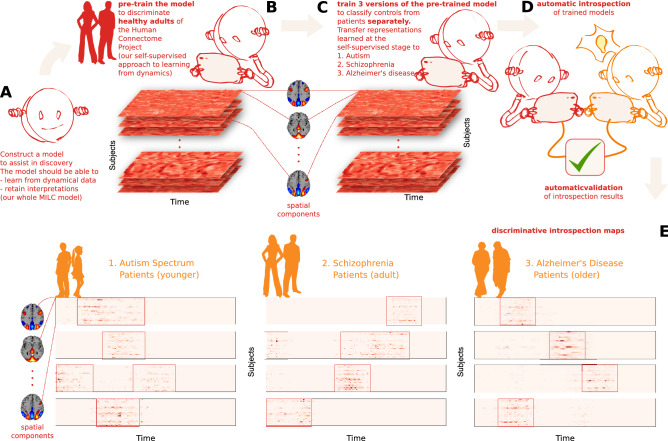
Figure 1.
An overview of our approach to model interpretation (created in program Inkscape 1.1.2, http://inkscape.org/release/inkscape-1.1.2). (A) Construct a model for disorder-specific discovery: we divided the entire ICA time courses into multiple sliding windows. Then we fed them into the whole MILC model that learns directly from the disorder signal dynamics and retains interpretations for further introspection. (B) Leverage self-supervised pretraining to distinguish healthy subjects: learned representations assist the model in maintaining its predictive power when downstream training data is limited. (C) Construct a downstream model to discriminate patients from controls for each disorder starting with the pre-trained whole MILC weights: transfer of representations learned during pretraining simplifies convergence and balances overfitting. (D) Introspection of the trained downstream models: we compute saliency maps as a rationale used by the model behind every prediction using interpretability methods to extract meaningful, distinctive parts of the data. Subsequently, the estimated salient aspects of the dynamics go through an automatic validation process. To this end, we use the most salient features to retrain an independent SML model that confirms the salience of the features. This information can then be relayed to a human expert in the relevant field to interpret further and advance knowledge about the disorders. (E) Examples of saliency maps as deemed highly predictive by the models for their predictions in three different discriminative tasks. Please note that the red boxes mark the highly discriminative salient parts of the data.