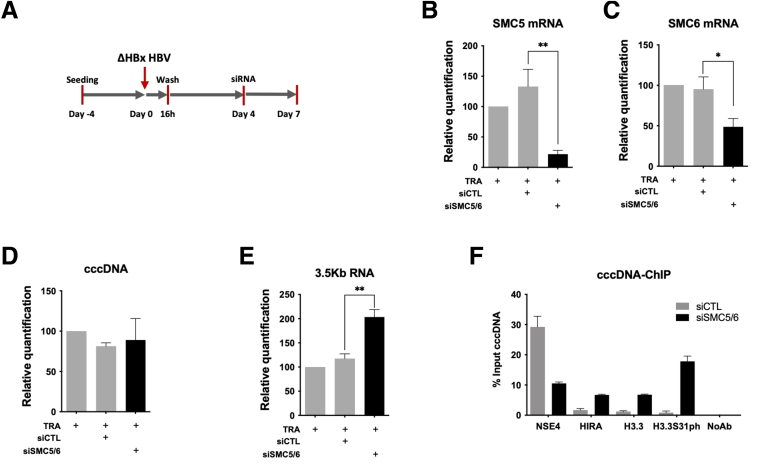
Figure 10.
ΔHBx-HBV transcription recovery is associated to enhanced HIRA and H3.3 recruitment to cccDNA. (A) Experimental timeline for SMC5/6 knock-down after HBV infection. HepG2hNTCP cells were infected for 16 hours and then extensively washed and transfected with siSMC5 and siSMC6 or siCTL at 4 dpi. Cells were harvested for analysis at 7 dpi. Messenger RNA (mRNA) expression of (B) SMC5 and (C) SMC6 after siRNA transfection was determined by real-time qPCR. (D) cccDNA and (E) 3.5-kb RNA amount was measured by qPCR at 7 dpi. cccDNA quantification was normalized over β-globin quantity, while relative 3.5-kb RNA amount was normalized over the housekeeping gene GUSb expression. (F) cccDNA-ChIP analysis using anti-NSE4, HIRA, H3.3, and H3.3S31ph antibodies at 9 dpi in CTL vs SMC5/6-depleted conditions. NoAb served as ChIP technical negative control. Graphs represent the means ± SEM of at least 3 independent experiments. The 2-tailed P value was calculated for a risk threshold of .05 using the 2/K sample permutation test with Monte Carlo resampling approximation. ∗P < .05 and ∗∗P < .01.