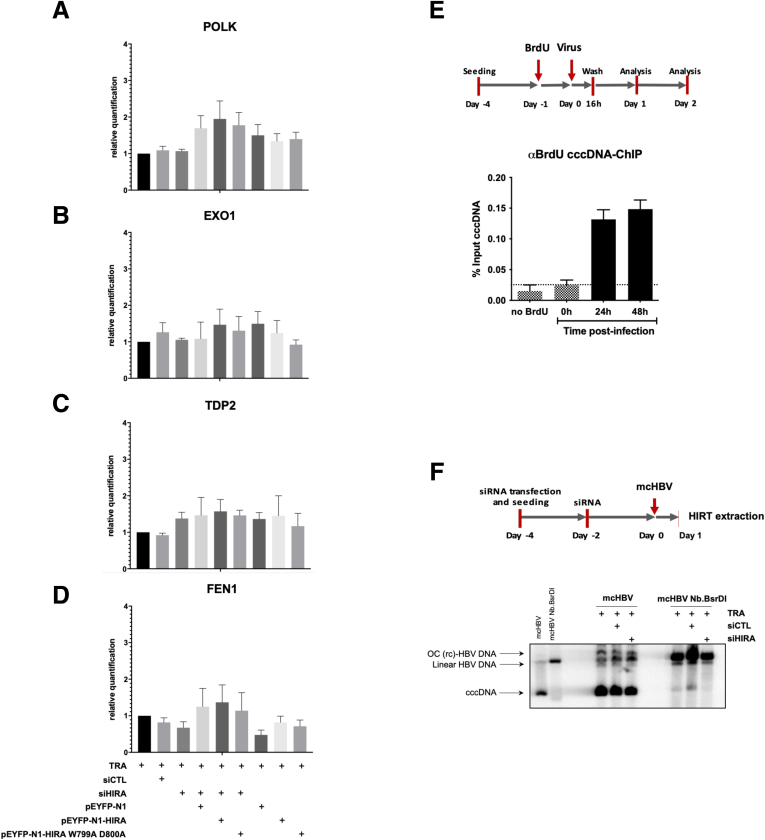
Figure 3.
Full cccDNA supercoiling in de novo infected cells occurs concomitantly to HBV genome repair. (A–D) Messenger RNA levels of (A) POLK, (B) EXO1, (C) TDP2, and (D) FEN1 were quantified by real-time qPCR assay and expressed as a percentage of TRA-treated cells after normalization over GUSb housekeeping gene expression. Data represent the means ± SEM of at least 3 independent experiments. (E) ChIP analysis of BrdU-containing cccDNA molecules. HepG2hNTCP cells were treated with 20 μmol/L for 24 hours before HBV infection and cultured for the indicated time points. cccDNA-ChIP qPCR using no antibody or anti-E2F antibody served as ChIP technical negative controls (Figure 8A and B), and the signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). Data are expressed as a percentage of enrichment with respect to initial input chromatin. (F) mcHBV constructs digested or not with Nb.BsrDI were transfected into HepG2hNTCP cells 48 hours after transfection with siHIRA or siCTL. Cells were harvested 24 hours after mcHBV transfection and the cccDNA amount was measured by Southern blot. Graphs represent the means ± SEM of at least 3 independent experiments. EXO1, Exonuclease 1; FEN-1, flap structure-specific endonuclease 1; OC (rc)-HBV DNA, open circular (relaxed circular) HBV DNA; POLK, DNA polymerase kappa; TDP2, tyrosyl-DNA phosphodiesterase-2.