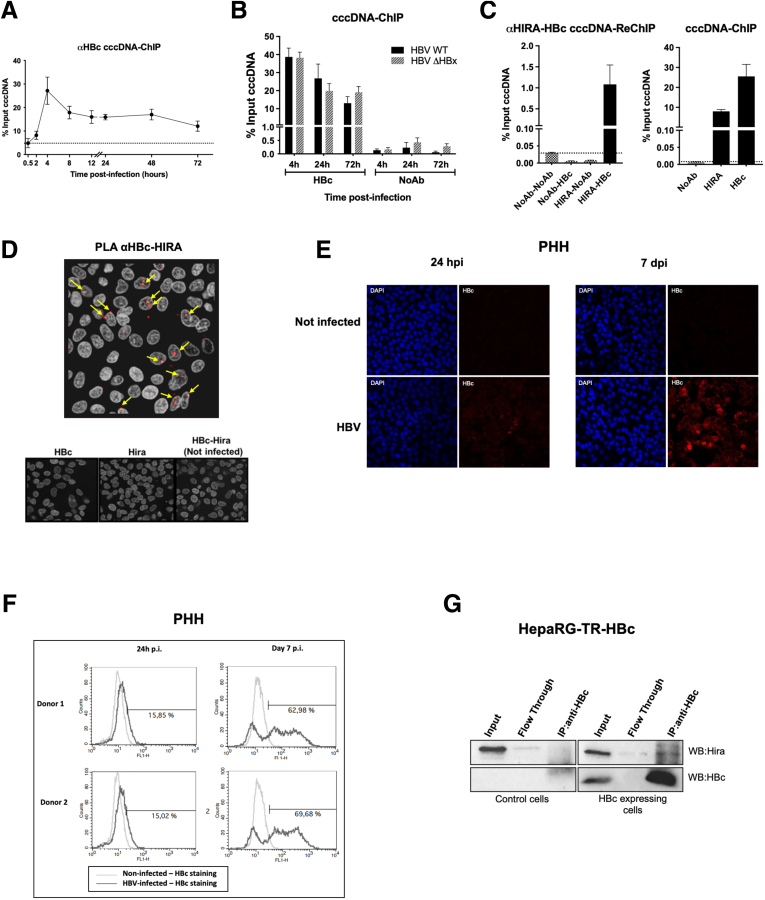
Figure 5.
Incoming HBV core protein associates to cccDNA and HIRA early after infection. (A and B) HepG2hNTCP cells were infected with either WT or ΔHBx HBV for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP analysis using an antibody against HBc. The levels of HBc on cccDNA were analyzed through the infection kinetics by ChIP-qPCR and expressed as the percentage of input chromatin. cccDNA-ChIP qPCR using no antibody (NoAb) or anti-E2F antibody served as technical negative controls (Figure 8A–D), and the signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). (C) the simultaneous presence of HIRA and HBc on the same cccDNA molecule was assessed by sequential ChIP-qPCR 24 hpi using an antibody against HIRA first and then an antibody directed against anti-HBc for immunoprecipitation. NoAb-NoAb, NoAb-HBc, and HIRA-NoAb combinations of sequential immunoprecipitation served as negative controls, and IP with single HBc and HIRA served as positive controls. Graphs represent the means ± SEM of at least 3 independent experiments. (D) Proximity between HBc and HIRA was assessed by PLA in HBV-infected PHHs at 24 hpi. The PLA signal is indicated by arrows. Uninfected and infected PHHs stained with only HBc or HIRA antibodies were used as negative controls (lower panels). (E) Immunofluorescence and (F) flow cytometry analysis of HBc-positive PHHs at 24 hpi and 7 dpi. Immunofluorescence was performed with antibody against HBc (red) and nuclei are stained by 4′,6-diamidino-2-phenylindole (DAPI) signal (blue). (G) Western blot analysis of HIRA-HBc immunoprecipitation in HepaRG cells inducible for HBc expression. Immunoprecipitation was performed with an anti-HBc antibody, using HepaRG-TR-HBc noninduced cells as a control (left panel). Western blot with anti-HIRA antibody showed a specific band in the immunoprecipitated fraction in the presence of HBc (right panel).