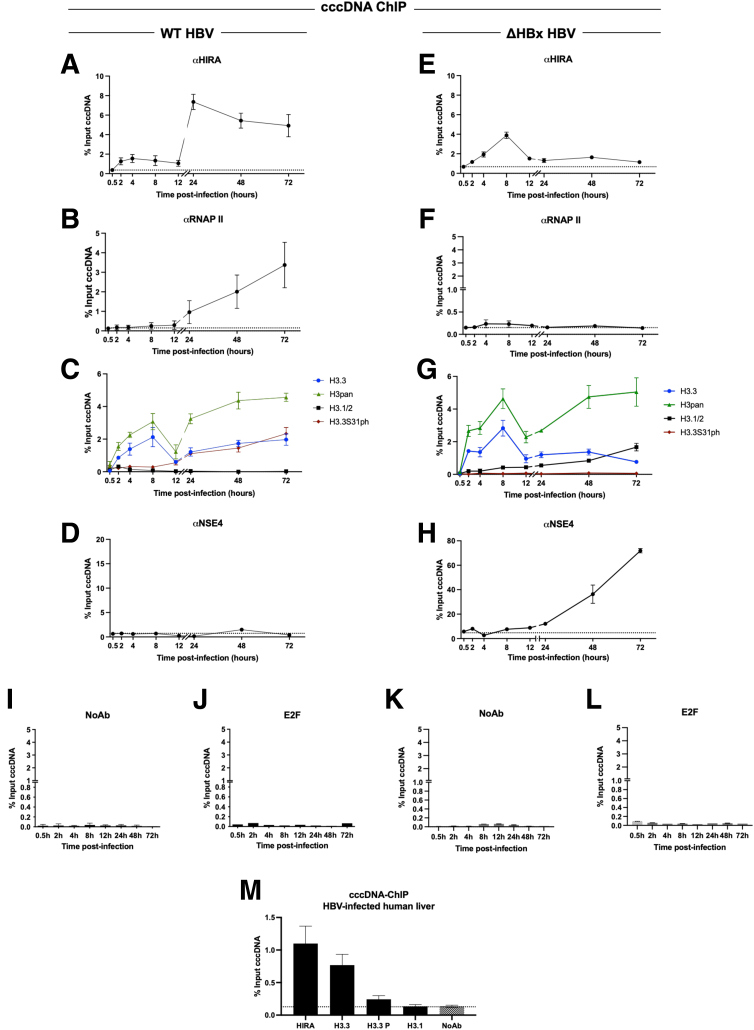
Figure 7.
Dynamics of HIRA and H3.3 recruitment and phosphorylation of H3.3S31 during de novo cccDNA formation and transcription of established cccDNA pool. HepG2hNTCP cells were infected with (A–D) WT or (E–H) ΔHBx-HBV for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP-qPCR analysis using antibodies against (A and E) histone chaperone HIRA, (B and F) RNAP II, (C and G) total H3 (H3pan) and histone variants H3.3 and H3.1/2 and H3.3S31ph, and (D and H) SMC5/6 complex subunit NSE4. (I–L) cccDNA-ChIP qPCR using no antibody (NoAb) or anti-E2F antibody served as technical negative controls. The signal at 0.5 hpi was considered a specific qPCR background for cccDNA quantification (Figure 1F). (M) Snap-frozen liver samples from 3 chronically infected male patients were subjected to cccDNA-ChIP with antibodies against HIRA, H3.3, H3.3S31ph, and H3.1/2. NoAb immunoprecipitation served as ChIP negative control. Data are expressed as a percentage of enrichment with respect to initial input chromatin and represent the means ± SEM of at least 3 independent experiments.