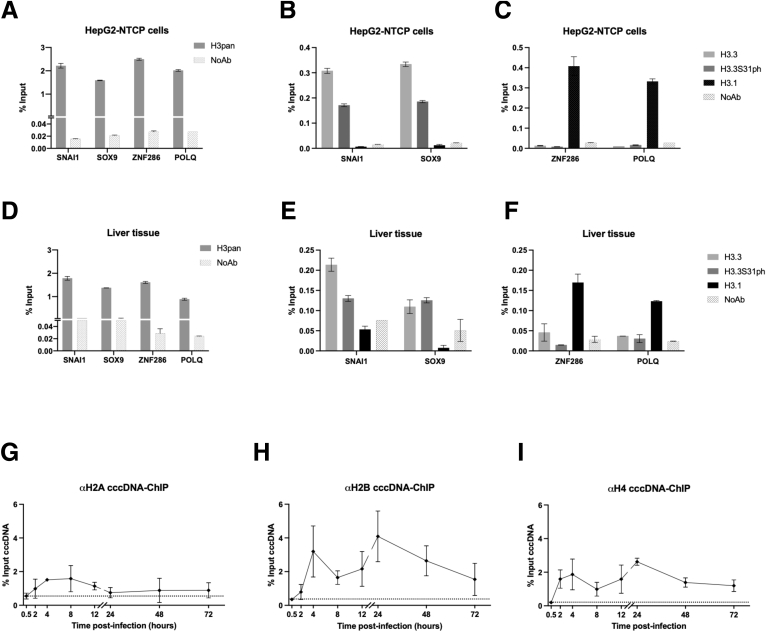
Figure 9.
Recruitment of the nucleosome core histones onto cccDNA. ChIP-qPCR analysis of genomic target sites shown to be preferentially bound by either H3.3 (SNAI1 and SOX9 promoters) or H3.1/2 (ZNF286 promoter and POLQ gene body) by using antibodies recognizing all H3 variants (H3pan), H3.3, H3.3S31ph, and H3.1/2 in (A–C) HBV-infected HepG2hNTCP cells at 72 hpi and (D–F) human liver tissue derived from 3 chronically HBV-infected patients. No antibody (NoAb) immuniprecipitation served as ChIP technical negative control. (G–I) HepG2hNTCP cells were infected at 250 viral genome equivalents/cell for up to 16 hours and then extensively washed and cultured for the indicated time points before ChIP-qPCR analysis using antibodies against histones H2A (G), H2B (H) and H4 (I). Data are expressed as a percentage of enrichment with respect to initial input chromatin and represent the means ± SEM of at least 3 independent experiments.