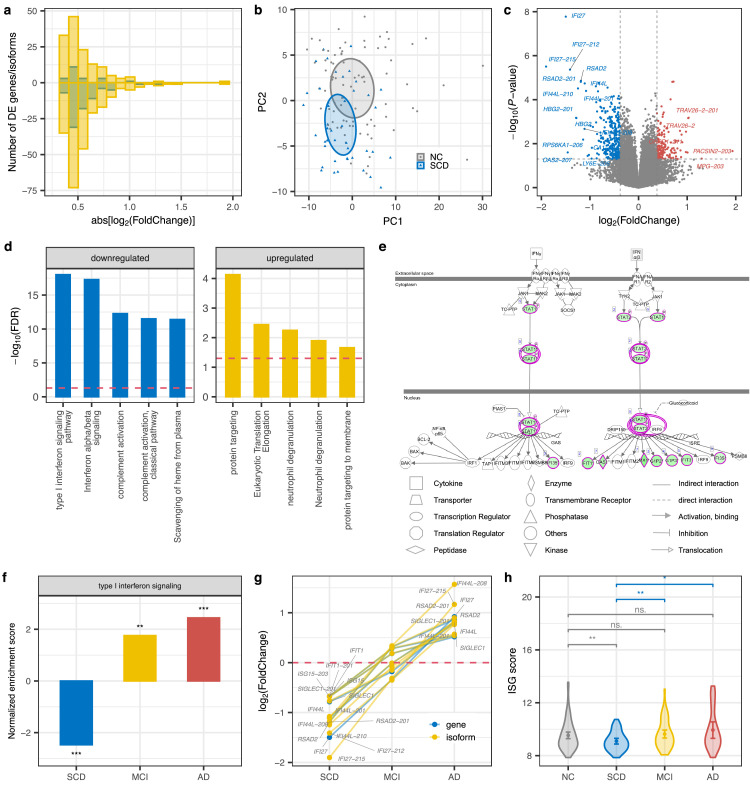
Figure 2.
Downregulation of the interferon signaling pathway in SCD. (a) Fold change (FC) histograms for up (positive number) or down-regulated (negative number) protein-coding genes (blue) and isoforms (yellow) in SCD. Abs: absolute. (b) PCA plot on differentially expressed genes and isoforms of SCD. (c) Volcano plot displaying the statistical significance (P-value) versus magnitude of change (fold change) of protein-coding genes and transcripts in SCD compared with NC. DE genes/isoforms with |log2(FC)| > 1 are labeled. (d) Pathway enrichment analysis for genes that were differentially expressed at gene or isoform level in SCD. The top 5 enriched pathways are shown. Blue and yellow represent down and up-regulated pathways, respectively. The red dashed line indicates the significance threshold (FDR = 0.05). (e) IPA canonical pathway for interferon signaling. Green shading represents down-regulated genes. (f) GSEA in type I interferon signaling pathways across disorders. Barplot shows the normalized enrichment score (NES) and significance. A positive/negative NES value indicates up/down-regulation of this gene set in disease compared with NC. ***, P < 0.001; **, P < 0.01. (g) Fold changes of ISG genes (IFI44L, IFI27, RSAD2, SIGLEC1, IFIT1, and IS15) defining the IFN signature at gene and isoform level across disorders. (h) ISG score of each group (NC = 82, SCD = 44, MCI = 51, AD = 25) based on the mean expression of these six signature genes. Error bar indicates 95% confidence interval of ISG score. P values were determined with the two-tailed t-test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ·, P < 0.1; ns., P > 0.1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)