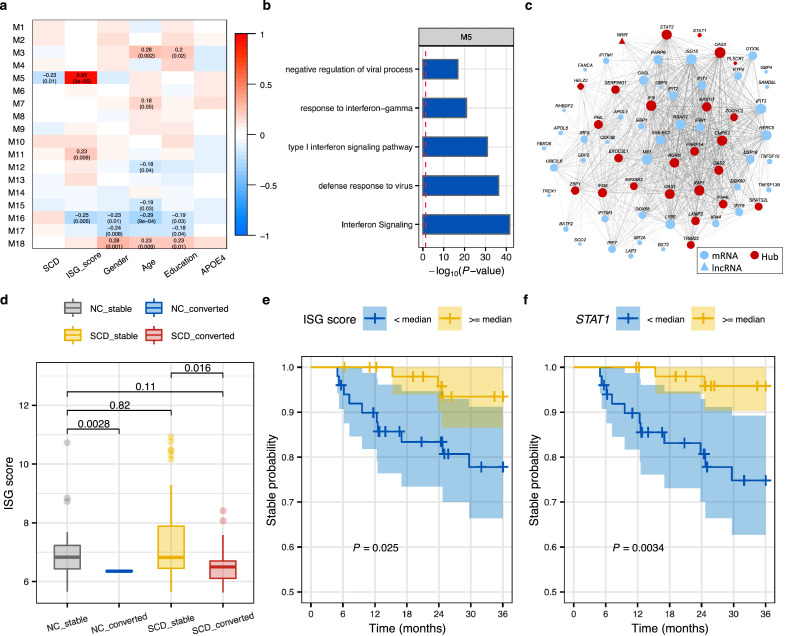
Figure 5.
Hub genes of the co-expression interferon signaling module serve as candidate biomarkers for conversion to MCI. (a) Pearson's correlation between module eigengenes and phenotypes. Left module-blocks represent the miRNA-lncRNA-mRNA co-expression modules at the gene level defined in SCD and NC samples. Correlation coefficients (top) and P-value (bottom) are shown in the grid, where red and blue colors indicate positive and negative correlations, respectively. Only significant associations (P < 0·05) are displayed. (b) Top 5 enriched (hypergeometric test) pathways of genes in the module M5. (c) The network of genes in the module M5, where only interaction edges with correlation coefficient > 0.15 are plotted. Edge thickness and node size correspond to pair-wise correlation and node connectivity, respectively. Node shapes represent different biotypes. Hub genes are highlighted with the red nodes. (d) The distribution of the ISG score in NC and SCD by conversion status (Stable NC = 24, converted NC = 2, Stable SCD = 88, converted SCD = 13). P values were determined with one-tailed t-test. (e-f) The Kaplan-Meier curves showing the 3-year disease conversion of SCD participants in ADNI (Alzheimer's Disease Neuroimaging Initiative) grouped by the median of the ISG score (e) and the expression of STAT1(f). Shadow indicates the 95% confidence interval. P-values of conversion difference were determined by Log-rank test. Yellow and blue denote higher or lower than the median level, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)