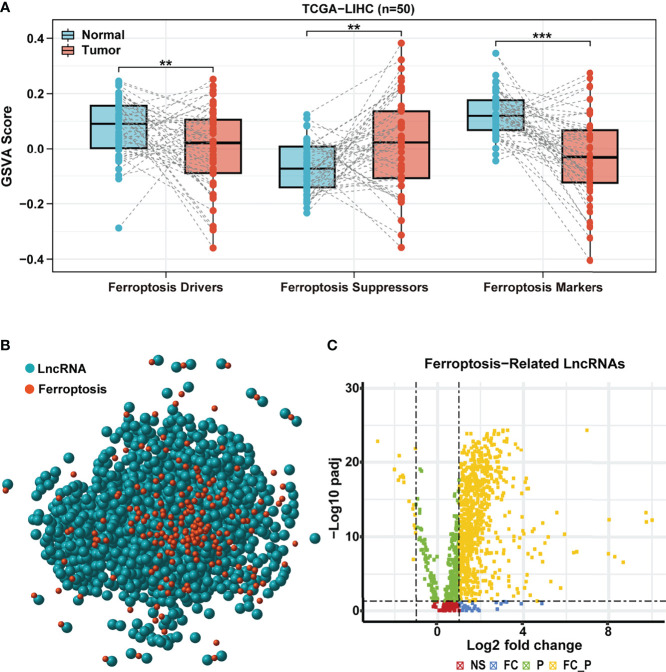
Figure 1.
Identification of differentially expressed ferroptosis-related lncRNA. (A) GSVA scores of ferroptosis drive, ferroptosis suppressor, and ferroptosis maker in paired samples of TCGA HCC cohort. (B) Correlation network of ferroptosis-related genes (red) and corresponding lncRNAs (blue). (C) The volcano plot for differentially expressed ferroptosis-related lncRNAs. Yellow dot represents significantly differentially expressed lncRNAs (|log2 fold-change| > 1 and FDR < 0.05). LncRNA, long non-coding RNA; GSVA, gene set variation analysis; TCGA, The Cancer Genome Atlas; HCC, hepatocellular carcinoma; FDR, false discovery rate.