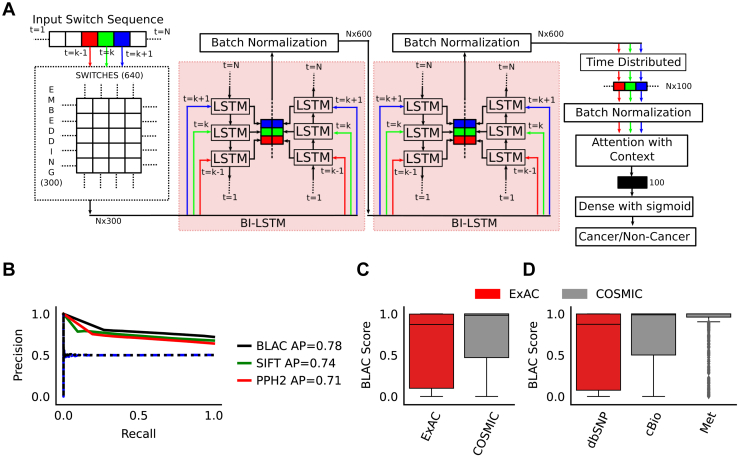
Figure 3.
Classification of cancerous and noncancerous variants.A, deep learning architecture, used for CRCS-based classification of ExAC/COSMIC variants. B, Precision-Recall (PR) curve for the BLAC after 200 epochs. The red and green curves indicate the performance of SIFT (20) and Polyphen2 (21), respectively. Validation performances were measured on fake alteration classes, constructed by randomly splitting cancer/noncancer alterations into two equal-size groups. The black dashed line represents the performance of the fake test set created from COSMIC data. Similarly, the blue dashed line is for ExAC data. Both PR curves thus obtained, as expected, collapsed on the 0.5 precision line. C, boxplots depict the distribution of prediction scores (probability of being a cancer alteration), assigned to the ExAC and COSMIC alterations, in the validation set (across all folds). D, similar trends are observed for nonpathogenic dbSNP alterations and mutations found in cancer patients from Met (23) and cBioPortal (62, 63). Scores on these datasets were predicted using the model trained on the full dataset. BLAC, bidirectional long short-term memory with attention & CRCS embeddings; COSMIC, Catalogue of Somatic Mutations In Cancer; CRCS, Continuous Representation of Codon Switches; ExAC, Exome Aggregation Consortium.