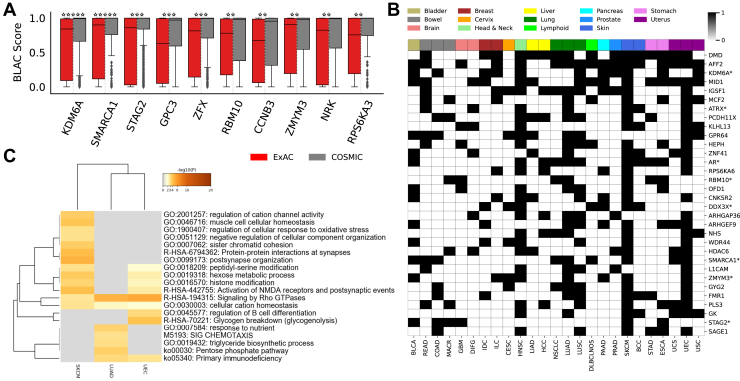
Figure 4.
Driver gene analysis and exploration.A, boxplots show the distribution of prediction scores assigned to ExAC and COSMIC alterations for the known driver genes from the validation set (across all folds). In the figure, five stars represent a p-value less than 5e−15. Values in the range [5e−15, 5e−12) are represented by four stars. Similarly, values in the range of [5e−12, 5e−9), [5e−9, 5e−6), and [5e−6, 5e−2) are represented by 3, 2, and 1 stars, respectively. B, heatmap shows the genes (in black) that have been marked significant most frequently, across cancer types. For a given cancer type in cBioPortal, a gene was marked significant if the BLAC scores of the reported mutations were significantly elevated as compared to dbSNP variants. The colors in the top row show the organ of cancer. Gene marked with ∗ are known driver genes. C, heatmap depicting the cluster-wise enrichment of the prominent biological functions in the indicated cancer types. Of note, the selected cancer types harbored a number of mutational genes identified using the CRCS-based approach. Cancer types that displayed significantly divergent risk groups include skin cutaneous melanoma (SKCM), lung adenocarcinoma (LUAD), and undifferentiated endometrial carcinoma (UEC). The scale bar represents the negatively log-transformed (base 10) p-values. BLAC, bidirectional long short-term memory with attention & CRCS embeddings; COSMIC, Catalogue of Somatic Mutations In Cancer; CRCS, Continuous Representation of Codon Switches; dbSNP, single nucleotide polymorphism database; ExAC, Exome Aggregation Consortium.