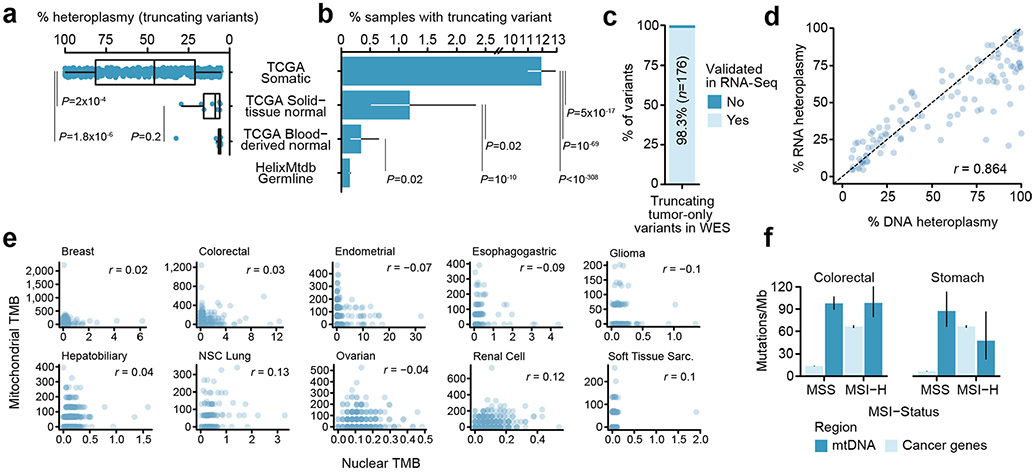
Extended Data Fig. 3 ∣. Analysis of mutation burden in normal tissues and of tumor mtDNA mutation burden with nuclear mutagenic processes.
a, Comparison of heteroplasmies between truncating variants detected in tumor tissue, adjacent normal tissue, and blood. P-values from two-sided Wilcoxon-rank sum test. Boxes are centered at the median and extended to from 25th-percentile to 75th-percentile; whiskers extend from 25/75th-percentiles to the largest value within 1.5 × IQR (interquartile range, 75th-percentile - 25th-percentile. b, Rate of truncating variants in TCGA tumors compared to matched non-malignant tissue, matched blood, and unmatched saliva samples from HelixMTdb. Truncating variants arise at 10-80-fold higher rate in tumors relative to normal tissues. Error bars are exact binomial 95% confidence intervals. P-values are from two-sided two-sample z-tests. c, The percentage of rescued truncating variants in TCGA that are recapitulated in orthogonal RNA sequencing from the same tumor sample. d, Correlation between heteroplasmies of rescued truncating variants in DNA and orthogonal RNA sequencing. Pearson correlation coefficient as shown. e, Mitochondrial and nuclear tumor mutation burdens (TMB, mutations/Mb) are shown for each well-covered tumor, among cancer types with n ≥ 100 samples. Nuclear TMBs are calculated based on mutations to 468 cancer-associated genes and their total exonic-sequence length. Pearson correlation coefficients r indicate no linear correlation between mitochondrial and nuclear TMBs were observed for any cancer type tested. f, TMBs for somatic mtDNA mutations and mutations to cancer-associated genes are compared between microsatellite stable (MSS) and microsatellite unstable (MSI-High) tumors, for both (n colorectal cancer: MSI=65, MSS=318; n stomach adenocarcinomas: MSI=75, MSS=256). Although MSI-High tumors have elevated TMB for nuclear cancer genes, there is no effect on mtDNA TMB. Moreover, mtDNA TMB is similar to (or exceeds) that of nuclear cancer associated genes in both cancer types. Error bars are 95% exact Poisson confidence intervals.