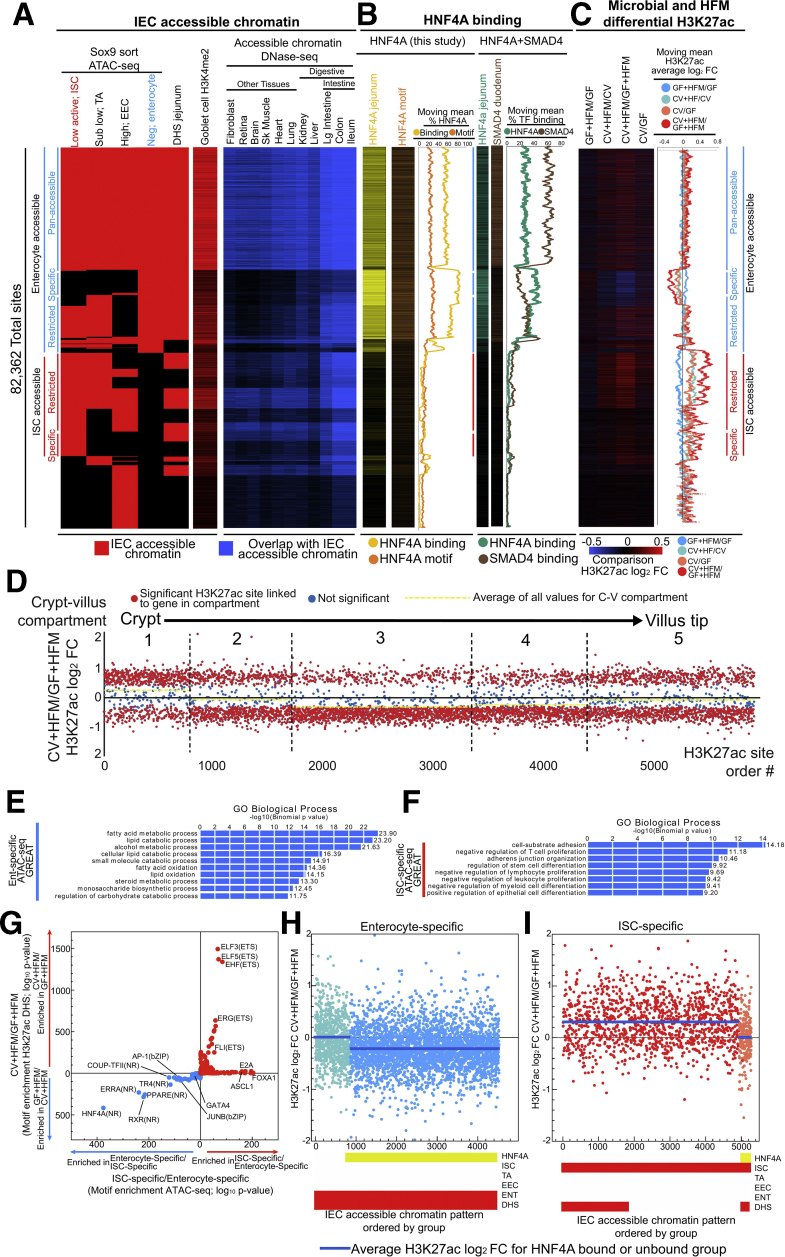
Figure 10.
HNF4A-dominated enterocyte and HNF4A-absent ISC regulatory regions behave distinctly in response to microbes and HFM. (A) Organized heatmap of merged accessible chromatin peak calls for sites that were significantly enriched in at least 1 of the 4 sorted IEC cell types (red).67 Jejunal DHS sites22 and H3K4me2 sites from goblet cells63 are also included. Groups of sites that are accessible in enterocytes and ISCs are further broken down into whether they are accessible in other (restricted) or only their IEC type (specific). A subset of sites are accessible across IEC subtypes (pan-accessible). Corresponding accessible chromatin data overlap with other mouse tissues (blue) identifies patterns of enrichment.21,68 Groups of sites that correspond to enterocyte sites are marked with a straight blue line, and ISC but not enterocytes sites are marked with straight red lines. (B) Heatmap for HNF4A binding (yellow) and computationally detected HNF4A motif (orange) for sites ordered as in (A). These data are summarized with descending moving means (500 site window, 1 site step). A similar pattern is seen using a previously published jejunum HNF4A ChIP-seq dataset (teal).54 Duodenum SMAD4 ChIP-seq dataset (brown).55 (C) Heatmap for H3K27ac differential binding for sites ordered as in (A). These data are summarized with descending moving means (500 site window, 1 site step). (D) The log2FC for H3K27ac sites that are significantly different in at least one comparison for the CV+HFM/GF+HFM comparison grouped by their linked neighboring gene into 1 of 5 compartments based on preferential expression along the crypt-villus axis.57 Within the compartments, H3K27ac sites are ordered by random. H3K27ac sites that are significant by the comparison on the Y-axis are red dots. All nonsignificant sites are blue. Yellow dashed lines are average for all sites within that compartment. (E) GREAT GO term enrichment for enterocyte-specific sites. (F) GREAT GO term enrichment for ISC-specific sites. (G) Motif enrichment at ATAC-seq sites linked to ISC-specific and Ent-specific groups versus CV+HFM and GF+HFM groups. For each group the reciprocal direction is used as the background (i.e., ISC-specific input versus Ent-specific background). The –log10P values are plotted. Both directions are plotted on the same axis with each analysis separated by colored arrows. (H) Groups of accessible sites (red bars) further organized by if they are bound by HNF4A (yellow bars) show dependence on HNF4A binding for decreased enrichment in CV+HFM/GF+HFM H3K27ac signal at enterocyte-specific sites and increased (I) ISC-specific sites on average.