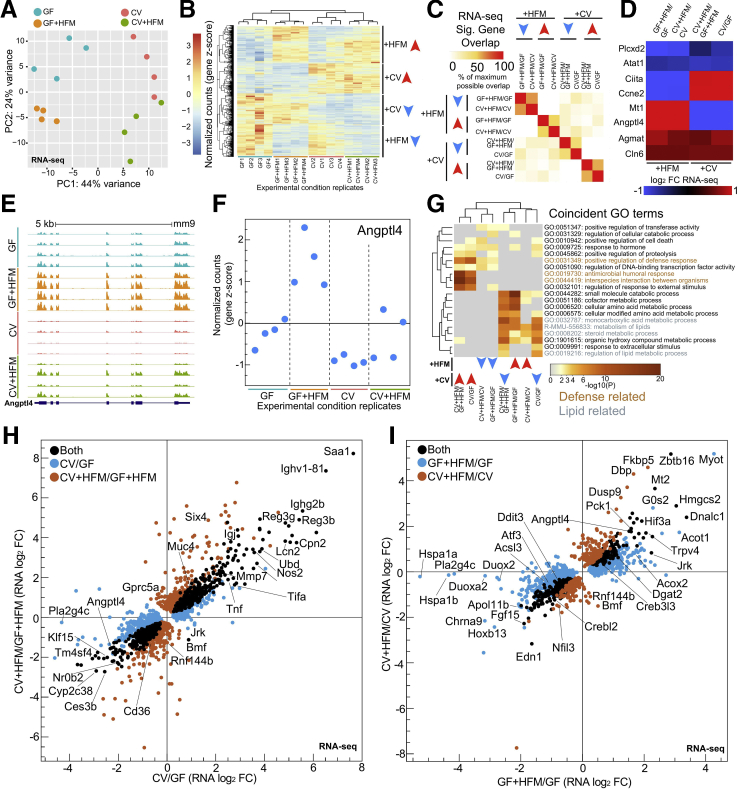
Figure 2.
Impact of HFM and microbes on gene transcription in the mammalian intestine. (A) DESeq2 PCA of RNA-seq normalized counts in each replicate for the 4 conditions, with 4 mice per condition. Adonis permutational multivariate ANOVA of RNA-seq distance matrix; microbes: P = .002, R2 = 0.240; meal: P = .005, R2 = 0.169. (B) Heatmap of row z-scored normalized counts for genes significantly differential in at least one comparison by RNA-seq (P adjusted < .05). Examples of blocks of commonly behaving genes are marked for +HFM and +CV directional groups. (C) Pairwise comparison of maximum overlap of coincident significant RNA-seq genes (P adjusted <.05) for 8 directional significance groups shows generally coincident directionality and genes for +CV and +HFM comparisons. (D) Heatmap of example genes significantly different in both a +CV and +HFM comparison (P adjusted <.05). (E) UCSC screenshot for RNA-seq replicate levels at the mouse Angptl4 locus. (F) RNA-seq z-scored normalized counts of Angptl4, which are significant in both +CV and +HFM comparisons, show amplified relative expression in the GF+HFM condition. (G) Clustered heatmap of significance values for shared GO terms in at least 2 of 8 directional RNA-seq significance groups. (H) Scatterplot of significantly different gene log2 fold change for CV/GF and CV+HFM/GF+HFM RNA-seq (P adjusted <.05). (I) Same as (H) for GF+HFM/GF and CV+HFM/CV (P adjusted <.05).