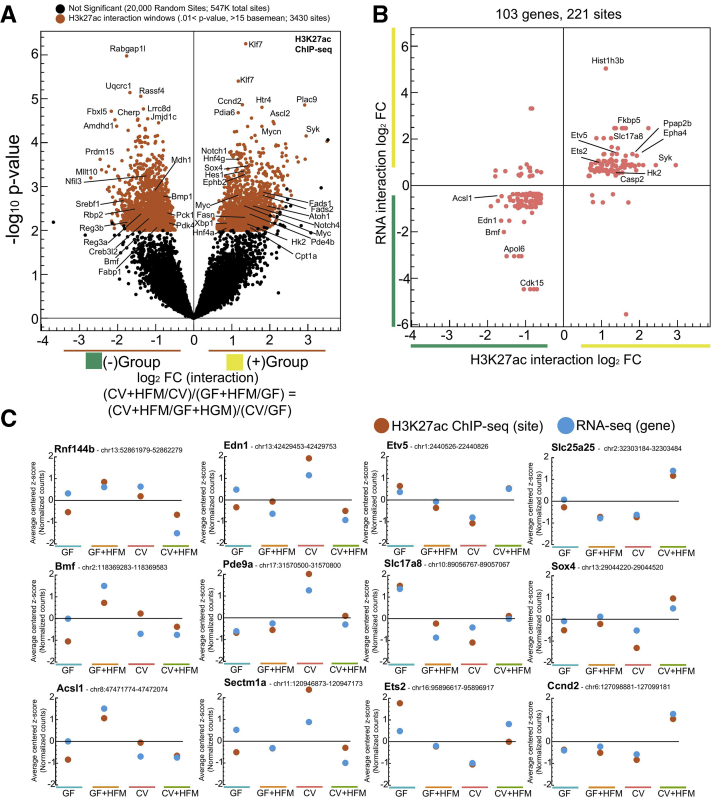
Figure 5.
Characterizing putative interaction regulatory regions. (A) Volcano plot showing interaction log2 fold change versus –log10P value for lenient (P value <.01, >15 base mean; red) cutoffs identifies H3K27ac regulatory windows with greatest potential for interaction. Twenty thousand out of 547,000+ enriched H3K27ac windows that did not pass the lenient interaction cutoff were chosen at random to represent noninteracting sites. (B) Scatterplot comparing H3K27ac log2 interaction windows fold change linked to the RNA log2 interaction fold change for genes that also show interaction reveals a positive correlation suggesting many of these regions are causal in contributing to the transcription patterns of these genes across +CV and +HFM conditions. (C) Selected putative H3K27ac interaction windows from each cluster showing consistent patterns of H3K27ac enrichment and relative RNA levels across the 4 conditions.