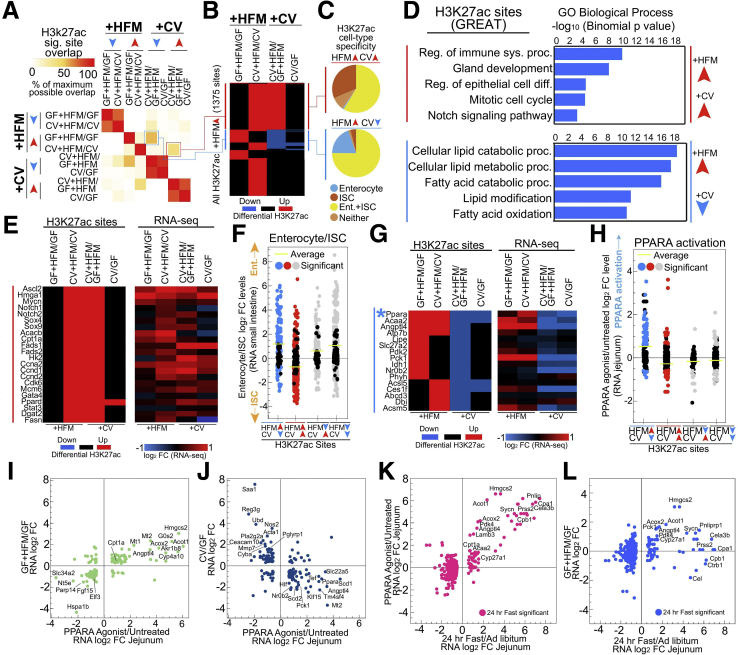
Figure 6.
Microbial and nutritional stimuli signal to many of the same intestinal regulatory regions. (A) Pairwise comparison of maximum overlap of coincident significant H3K27ac sites (P adjusted <.05) for 8 directional significance groups identifies microbially responsive regulatory regions with CV+HFM/CV-up also being CV+HFM/GF+HFM-up (red) and GF+HFM/GF-up also being CV+HFM/GF+HFM-down (blue). (B) Heatmap of differential comparisons (P adjusted <.05) for all +HFM-up sites shows the proportion that is also microbially responsive. (C) Pie charts for red and blue +HFM-up groups that show the proportion that overlap with previously characterized enterocyte and ISC regulatory regions.45 (D) GREAT GO term enrichment for red and blue +HFM-up H3K27ac groups. (E) Heatmap of example red +HFM-up and +CV-up H3K27ac sites and their linked gene’s RNA-seq log2 fold change, including many loci associated with ISCs and proliferation. (F) Different combinations of differential H3K27ac sites that are both +HFM and +CV responsive show that only red sites that are +HFM-up and +CV-up are linked to genes that are preferentially expressed in ISCs relative to enterocytes. (G) Heatmap of example blue +HFM-up H3K27ac sites and their linked gene’s RNA-seq log2 fold change. Blue asterisk marks Ppara regulatory region that is characterized in Figure 7. (H) Different combinations of differential H3K27ac sites that are both +HFM and +CV responsive show that only blue sites that are +HFM-up and +CV-down are linked to genes that are activated by PPARA. (I) Scatterplot of genes that are significantly differential after PPARA activation and in GF+HFM/GF show a positive correlation. (J) Scatterplot of genes that are significantly differential after PPARA activation and in CV/GF show a negative correlation. (K) Scatterplot of log2 fold change levels for genes significant (<.005 P adjusted) in 24-hour fast/ad libitum versus PPARA activated genes.48 (L) Scatterplot of log2 fold change levels for genes significant (<.005 P adjusted) in 24-hour fast/ad libitum versus GF+HFM/GF shows many of the same PPARA targets are activated by fasting and HFM.48