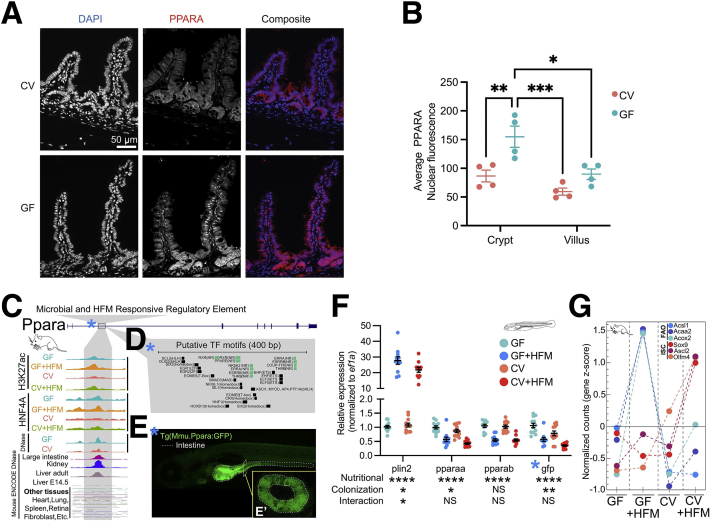
Figure 7.
Characterization of the putative PPARA regulatory region that responds to microbes and HFM. (A) PPARA immunofluorescence of small intestinal villi (red) in GF and CV mice fed ad libitum. (B) Quantification of IEC PPARA nuclear fluorescence for the crypt and villus identifies higher nuclear fluorescence in GF mice. ∗P value ≤.05, ∗∗P value ≤.01, and ∗∗∗P value ≤.001; two-way ANOVA; n = 4 per condition. (C) Exploded view of H3K27ac region at mouse Ppara locus from Figure 6G that is +HFM-up and +CV-down showing signal across conditions for H3K27ac, HNF4A, and accessible chromatin for jejunum and numerous other tissues. (D) Putative TF motifs at the mouse Ppara regulatory region include multiple nuclear receptor sites, including PPARE. (E) Transgenic Tg(Mmu.Ppara:GFP) zebrafish at 6 days post-fertilization (dpf) with the mouse Ppara regulatory region upstream of a mouse cFos minimal promoter driving GFP shows expression largely limited to the anterior intestine. E' boxed inset shows a confocal cross section confirming the signal is specific to IECs. (F) Quantitative real-time polymerase chain reaction of a 4 condition experiment using whole 6 dpf Tg(Mmu.Ppara:GFP) zebrafish shows similar responses to colonization and HFM for pparaa and gfp. Significance calls for colonization, nutritional, and interaction based on 2-factor ANOVA: ∗P value ≤.05, ∗∗P value ≤.01, and ∗∗∗∗P value ≤.0001. Ten to 20 larvae per replicates; 11-12 replicates per condition. (G) FAO and ISC associated genes showing particular expression in IECs from GF+HFM and CV+HFM mice, respectively (Supplementary Table 2).