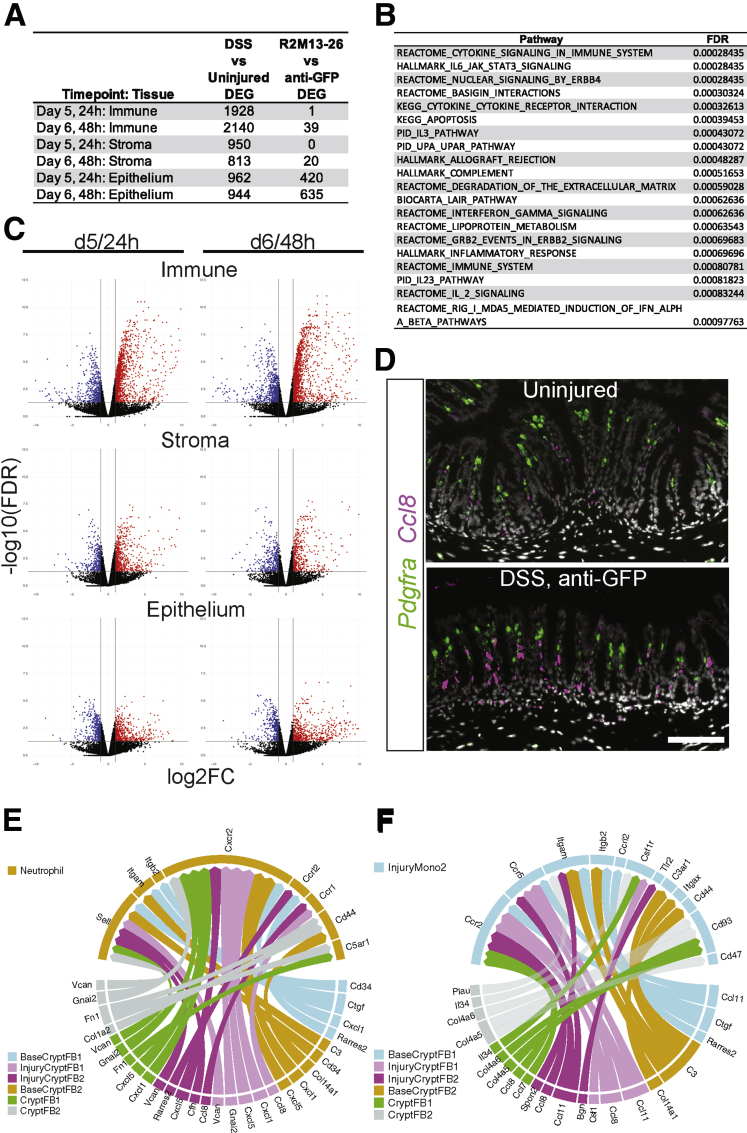
Figure 10.
DSS damage induced large-scale inflammatory responses, including potential stromal to immune cell signaling. (A) The number of genes that are significantly differentially expressed (false discovery rate [FDR] < 0.05, log2FC ≥ |1|) in each tissue layer or lineage by time point in 1 of the 2 comparisons: DSS damage (anti-GFP) vs uninjured and SZN-1326-p/DSS damage vs anti-GFP/DSS damage. (B) The top 20 gene sets or pathways identified by GSEA that were enriched in the stromal cells upon DSS treatment compared with the uninjured state at day 5. (C) Volcano plots of the differentially expressed genes between the DSS-damage and uninjured conditions for the indicated tissue lineage or layer and time point. Genes that display at least a 2-fold change and have an FDR of <0.05 are indicated in red (increased) and blue (decreased). Any gene with an FDR lower than 10E-12 was floored at 10E-12. (D) RNA in situ hybridization of the chemokine Ccl8 and the fibroblast marker Pdgfra in the colon of uninjured, and DSS/anti-GFP samples at day 5. Scale bar represents 100 mm. (E, F) Cell-cell interaction plots displaying the top 5% of potential signaling interactions between the cell types represented in the bottom half of the circle with the target cell type represented by the top half of the circle. The arrows represent the ligand toward the receptor.